PURA syndrome-causing mutations impair PUR-domain integrity and affect P-body association
- PMID: 38655849
- PMCID: PMC11042805
- DOI: 10.7554/eLife.93561
PURA syndrome-causing mutations impair PUR-domain integrity and affect P-body association
Abstract
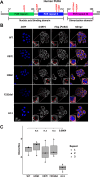
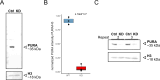
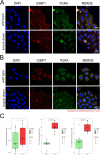
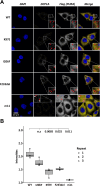
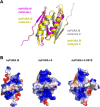
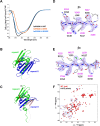
Mutations in the human PURA gene cause the neurodevelopmental PURA syndrome. In contrast to several other monogenetic disorders, almost all reported mutations in this nucleic acid-binding protein result in the full disease penetrance. In this study, we observed that patient mutations across PURA impair its previously reported co-localization with processing bodies. These mutations either destroyed the folding integrity, RNA binding, or dimerization of PURA. We also solved the crystal structures of the N- and C-terminal PUR domains of human PURA and combined them with molecular dynamics simulations and nuclear magnetic resonance measurements. The observed unusually high dynamics and structural promiscuity of PURA indicated that this protein is particularly susceptible to mutations impairing its structural integrity. It offers an explanation why even conservative mutations across PURA result in the full penetrance of symptoms in patients with PURA syndrome.
Keywords: PURA; PURA syndrome; RNA binding; X-ray crystallography; cell biology; human; molecular biophysics; protein folding stability; structural biology.
Plain language summary
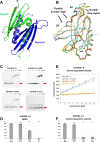
PURA syndrome is a neurodevelopmental disorder that affects about 650 patients worldwide, resulting in a range of symptoms including neurodevelopmental delays, intellectual disability, muscle weakness, seizures, and eating difficulties. The condition is caused by a mutated gene that codes for a protein called PURA. PURA binds RNA – the molecule that carries genetic information so it can be translated into proteins – and has roles in regulating the production of new proteins. Contrary to other conditions that result from mutations in a single gene, PURA syndrome patients show ‘high penetrance’, meaning almost every reported mutation in the gene leads to symptoms. Proske, Janowski et al. wanted to understand the molecular basis for this high penetrance. To find out more, the researchers first examined how patient mutations affected the location of the PURA in the cell, using human cells grown in the laboratory. Normally, PURA travels to P-bodies, which are groupings of RNA and proteins involved in regulating which genes get translated into proteins. The researchers found that in cells carrying PURA syndrome mutations, PURA failed to move adequately to P-bodies. To find out how this ‘mislocalization’ might happen, Proske, Janowski et al. tested how different mutations affected the three-dimensional folding of PURA. These analyses showed that the mutations impair the protein’s folding and thereby disrupt PURA’s ability to bind RNA, which may explain why mutant PURA cannot localize correctly. Proske, Janowski et al. describe the molecular abnormalities of PURA underlying this disorder and show how molecular analysis of patient mutations can reveal the mechanisms of a disease at the cell level. The results show that the impact of mutations on the structural integrity of the protein, which affects its ability to bind RNA, are likely key to the symptoms of the syndrome. Additionally, their approach used establishes a way to predict and test mutations that will cause PURA syndrome. This may help to develop diagnostic tools for this condition.
© 2024, Proske, Janowski et al.
Conflict of interest statement
MP, RJ, SB, HK, TM, TK, SH, JT, AC, LM, AV, RF, ED, GD, DD, MS, DN No competing interests declared
Figures











Update of
- doi: 10.1101/2023.09.19.558386
- doi: 10.7554/eLife.93561.1
- doi: 10.7554/eLife.93561.2
Similar articles
-
The Molecular Function of PURA and Its Implications in Neurological Diseases.Front Genet. 2021 Mar 11;12:638217. doi: 10.3389/fgene.2021.638217. eCollection 2021. Front Genet. 2021. PMID: 33777106 Free PMC article. Review.
-
PURA, the gene encoding Pur-alpha, member of an ancient nucleic acid-binding protein family with mammalian neurological functions.Gene. 2018 Feb 15;643:133-143. doi: 10.1016/j.gene.2017.12.004. Epub 2017 Dec 6. Gene. 2018. PMID: 29221753 Free PMC article. Review.
-
PURA syndrome: clinical delineation and genotype-phenotype study in 32 individuals with review of published literature.J Med Genet. 2018 Feb;55(2):104-113. doi: 10.1136/jmedgenet-2017-104946. Epub 2017 Nov 2. J Med Genet. 2018. PMID: 29097605 Free PMC article. Review.
-
Depletion of the RNA-binding protein PURA triggers changes in posttranscriptional gene regulation and loss of P-bodies.Nucleic Acids Res. 2023 Feb 22;51(3):1297-1316. doi: 10.1093/nar/gkac1237. Nucleic Acids Res. 2023. PMID: 36651277 Free PMC article.
-
Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability.J Med Genet. 2014 Dec;51(12):806-13. doi: 10.1136/jmedgenet-2014-102798. Epub 2014 Oct 23. J Med Genet. 2014. PMID: 25342064 Free PMC article.
Cited by
-
Purine-Rich Element Binding Protein Alpha, a Nuclear Matrix Protein, Has a Role in Prostate Cancer Progression.Int J Mol Sci. 2024 Jun 24;25(13):6911. doi: 10.3390/ijms25136911. Int J Mol Sci. 2024. PMID: 39000020 Free PMC article. Review.
-
PURA syndrome-a genetic cause of a neurodevelopmental disorder-case report.Front Pediatr. 2025 Jul 10;13:1607213. doi: 10.3389/fped.2025.1607213. eCollection 2025. Front Pediatr. 2025. PMID: 40708900 Free PMC article.
-
Inferring gene regulatory networks of ALS from blood transcriptome profiles.Heliyon. 2024 Nov 26;10(23):e40696. doi: 10.1016/j.heliyon.2024.e40696. eCollection 2024 Dec 15. Heliyon. 2024. PMID: 39687198 Free PMC article.
-
Inherited PURA Pathogenic Variant Associated With a Mild Neurodevelopmental Disorder.Neurol Genet. 2024 Aug 6;10(5):e200181. doi: 10.1212/NXG.0000000000200181. eCollection 2024 Oct. Neurol Genet. 2024. PMID: 39131487 Free PMC article.
-
PURA-Related Neurodevelopmental Disorders with Epilepsy Treated with Ketogenic Diet: A Case-Based Review.Genes (Basel). 2024 Jun 27;15(7):848. doi: 10.3390/genes15070848. Genes (Basel). 2024. PMID: 39062627 Free PMC article. Review.
References
-
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

