Construction of the ETECFinder database for the characterization of enterotoxigenic Escherichia coli (ETEC) and revision of the VirulenceFinder web tool at the CGE website
- PMID: 38656142
- PMCID: PMC11237473
- DOI: 10.1128/jcm.00570-23
Construction of the ETECFinder database for the characterization of enterotoxigenic Escherichia coli (ETEC) and revision of the VirulenceFinder web tool at the CGE website
Abstract
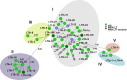
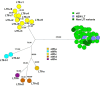
The identification of pathogens is essential for effective surveillance and outbreak detection, which lately has been facilitated by the decreasing cost of whole-genome sequencing (WGS). However, extracting relevant virulence genes from WGS data remains a challenge. In this study, we developed a web-based tool to predict virulence-associated genes in enterotoxigenic Escherichia coli (ETEC), which is a major concern for human and animal health. The database includes genes encoding the heat-labile toxin (LT) (eltA and eltB), heat-stable toxin (ST) (est), colonization factors CS1 through 30, F4, F5, F6, F17, F18, and F41, as well as toxigenic invasion and adherence loci (tia, tibAC, etpBAC, eatA, yghJ, and tleA). To construct the database, we revised the existing ETEC nomenclature and used the VirulenceFinder webtool at the CGE website [VirulenceFinder 2.0 (dtu.dk)]. The database was tested on 1,083 preassembled ETEC genomes, two BioProjects (PRJNA421191 with 305 and PRJNA416134 with 134 sequences), and the ETEC reference genome H10407. In total, 455 new virulence gene alleles were added, 50 alleles were replaced or renamed, and two were removed. Overall, our tool has the potential to greatly facilitate ETEC identification and improve the accuracy of WGS analysis. It can also help identify potential new virulence genes in ETEC. The revised nomenclature and expanded gene repertoire provide a better understanding of the genetic diversity of ETEC. Additionally, the user-friendly interface makes it accessible to users with limited bioinformatics experience.
Importance: Detecting colonization factors in enterotoxigenic Escherichia coli (ETEC) is challenging due to their large number, heterogeneity, and lack of standardized tests. Therefore, it is important to include these ETEC-related genes in a more comprehensive VirulenceFinder database in order to obtain a more complete coverage of the virulence gene repertoire of pathogenic types of E. coli. ETEC vaccines are of great importance due to the severity of the infections, primarily in children. A tool such as this could assist in the surveillance of ETEC in order to determine the prevalence of relevant types in different parts of the world, allowing vaccine developers to target the most prevalent types and, thus, a more effective vaccine.
Keywords: CGE website; ETEC virulence genes; WGS tool; curated database; enterotoxigenic E. coli.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, Praet N, Bellinger DC, de Silva NR, Gargouri N, Speybroeck N, Cawthorne A, Mathers C, Stein C, Angulo FJ, Devleesschauwer B, World Health Organization Foodborne Disease Burden Epidemiology Reference Group . 2015. World health organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med 12:e1001923. doi: 10.1371/journal.pmed.1001923 - DOI - PMC - PubMed
-
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, et al. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2 - DOI - PubMed
-
- Murphy D, Ricci A, Auce Z, Beechinor JG, Bergendahl H, Breathnach R, Bureš J, Duarte Da Silva JP, Hederová J, Hekman P, et al. 2017. EMA and EFSA joint scientific opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety (RONAFA). EFSA J 15:e04666. doi: 10.2903/j.efsa.2017.4666 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

