Japanese encephalitis virus NS1 and NS1' proteins induce vimentin rearrangement via the CDK1-PLK1 axis to promote viral replication
- PMID: 38656209
- PMCID: PMC11092344
- DOI: 10.1128/jvi.00195-24
Japanese encephalitis virus NS1 and NS1' proteins induce vimentin rearrangement via the CDK1-PLK1 axis to promote viral replication
Abstract
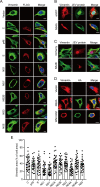
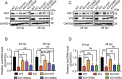
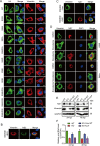
The host cytoskeleton plays crucial roles in various stages of virus infection, including viral entry, transport, replication, and release. However, the specific mechanisms by which intermediate filaments are involved in orthoflavivirus infection have not been well understood. In this study, we demonstrate that the Japanese encephalitis virus (JEV) remodels the vimentin network, resulting in the formation of cage-like structures that support viral replication. Mechanistically, JEV NS1 and NS1' proteins induce the translocation of CDK1 from the nucleus to the cytoplasm and interact with it, leading to the phosphorylation of vimentin at Ser56. This phosphorylation event recruits PLK1, which further phosphorylates vimentin at Ser83. Consequently, these phosphorylation modifications convert the typically filamentous vimentin into non-filamentous "particles" or "squiggles." These vimentin "particles" or "squiggles" are then transported retrogradely along microtubules to the endoplasmic reticulum, where they form cage-like structures. Notably, NS1' is more effective than NS1 in triggering the CDK1-PLK1 cascade response. Overall, our study provides new insights into how JEV NS1 and NS1' proteins manipulate the vimentin network to facilitate efficient viral replication.
Importance: Japanese encephalitis virus (JEV) is a mosquito-borne orthoflavivirus that causes severe encephalitis in humans, particularly in Asia. Despite the availability of a safe and effective vaccine, JEV infection remains a significant public health threat due to limited vaccination coverage. Understanding the interactions between JEV and host proteins is essential for developing more effective antiviral strategies. In this study, we investigated the role of vimentin, an intermediate filament protein, in JEV replication. Our findings reveal that JEV NS1 and NS1' proteins induce vimentin rearrangement, resulting in the formation of cage-like structures that envelop the viral replication factories (RFs), thus facilitating efficient viral replication. Our research highlights the importance of the interplay between the cytoskeleton and orthoflavivirus, suggesting that targeting vimentin could be a promising approach for the development of antiviral strategies to inhibit JEV propagation.
Keywords: CDK1; Japanese encephalitis virus; PLK1; infection; vimentin rearrangement.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

