Dependence of nucleosome mechanical stability on DNA mismatches
- PMID: 38656237
- PMCID: PMC11042804
- DOI: 10.7554/eLife.95514
Dependence of nucleosome mechanical stability on DNA mismatches
Abstract
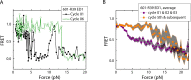
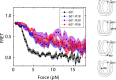
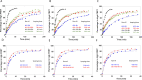
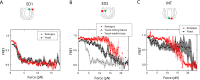
The organization of nucleosomes into chromatin and their accessibility are shaped by local DNA mechanics. Conversely, nucleosome positions shape genetic variations, which may originate from mismatches during replication and chemical modification of DNA. To investigate how DNA mismatches affect the mechanical stability and the exposure of nucleosomal DNA, we used an optical trap combined with single-molecule FRET and a single-molecule FRET cyclization assay. We found that a single base-pair C-C mismatch enhances DNA bendability and nucleosome mechanical stability for the 601-nucleosome positioning sequence. An increase in force required for DNA unwrapping from the histone core is observed for single base-pair C-C mismatches placed at three tested positions: at the inner turn, at the outer turn, or at the junction of the inner and outer turn of the nucleosome. The results support a model where nucleosomal DNA accessibility is reduced by mismatches, potentially explaining the preferred accumulation of single-nucleotide substitutions in the nucleosome core and serving as the source of genetic variation during evolution and cancer progression. Mechanical stability of an intact nucleosome, that is mismatch-free, is also dependent on the species as we find that yeast nucleosomes are mechanically less stable and more symmetrical in the outer turn unwrapping compared to Xenopus nucleosomes.
Keywords: DNA mismatch; DNA repair; S. cerevisiae; fluorescence resonance energy transfer; molecular biophysics; nucleosome; optical tweezers; single molecule biophysics; structural biology; xenopus.
© 2024, Ngo et al.
Conflict of interest statement
TN, BL, FW, AB, CW, TH No competing interests declared
Figures





Update of
- doi: 10.1101/2022.11.21.517409
- doi: 10.7554/eLife.95514.1
- doi: 10.7554/eLife.95514.2
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

