Crystal structure of human peptidylarginine deiminase type VI (PAD6) provides insights into its inactivity
- PMID: 38656308
- PMCID: PMC11067741
- DOI: 10.1107/S2052252524002549
Crystal structure of human peptidylarginine deiminase type VI (PAD6) provides insights into its inactivity
Abstract
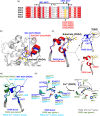
Human peptidylarginine deiminase isoform VI (PAD6), which is predominantly limited to cytoplasmic lattices in the mammalian oocytes in ovarian tissue, is essential for female fertility. It belongs to the peptidylarginine deiminase (PAD) enzyme family that catalyzes the conversion of arginine residues to citrulline in proteins. In contrast to other members of the family, recombinant PAD6 was previously found to be catalytically inactive. We sought to provide structural insight into the human homologue to shed light on this observation. We report here the first crystal structure of PAD6, determined at 1.7 Å resolution. PAD6 follows the same domain organization as other structurally known PAD isoenzymes. Further structural analysis and size-exclusion chromatography show that PAD6 behaves as a homodimer similar to PAD4. Differential scanning fluorimetry suggests that PAD6 does not coordinate Ca2+ which agrees with acidic residues found to coordinate Ca2+ in other PAD homologs not being conserved in PAD6. The crystal structure of PAD6 shows similarities with the inactive state of apo PAD2, in which the active site conformation is unsuitable for catalytic citrullination. The putative active site of PAD6 adopts a non-productive conformation that would not allow protein-substrate binding due to steric hindrance with rigid secondary structure elements. This observation is further supported by the lack of activity on the histone H3 and cytokeratin 5 substrates. These findings suggest a different mechanism for enzymatic activation compared with other PADs; alternatively, PAD6 may exert a non-enzymatic function in the cytoplasmic lattice of oocytes and early embryos.
Keywords: PAD6; cytoplasmic lattices; human peptidylarginine deiminase VI; mammalian fertilization; protein structures.
open access.
Figures



References
-
- Arita, K., Hashimoto, H., Shimizu, T., Nakashima, K., Yamada, M. & Sato, M. (2004). Nat. Struct. Mol. Biol. 11, 777–783. - PubMed
-
- Begemann, M., Rezwan, F. I., Beygo, J., Docherty, L. E., Kolarova, J., Schroeder, C., Buiting, K., Chokkalingam, K., Degenhardt, F., Wakeling, E. L., Kleinle, S., González Fassrainer, D., Oehl-Jaschkowitz, B., Turner, C. L. S., Patalan, M., Gizewska, M., Binder, G., Bich Ngoc, C. T., Chi Dung, V., Mehta, S. G., Baynam, G., Hamilton-Shield, J. P., Aljareh, S., Lokulo-Sodipe, O., Horton, R., Siebert, R., Elbracht, M., Temple, I. K., Eggermann, T. & Mackay, D. J. G. (2018). J. Med. Genet. 55, 497–504. - PMC - PubMed
-
- Boggs, J. M., Rangaraj, G., Koshy, K. M., Ackerley, C., Wood, D. D. & Moscarello, M. A. (1999). J. Neurosci. Res. 57, 529–535. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

