Chaperone-mediated MHC-I peptide exchange in antigen presentation
- PMID: 38656309
- PMCID: PMC11067752
- DOI: 10.1107/S2052252524002768
Chaperone-mediated MHC-I peptide exchange in antigen presentation
Abstract
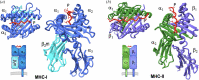
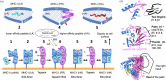
This work focuses on molecules that are encoded by the major histocompatibility complex (MHC) and that bind self-, foreign- or tumor-derived peptides and display these at the cell surface for recognition by receptors on T lymphocytes (T cell receptors, TCR) and natural killer (NK) cells. The past few decades have accumulated a vast knowledge base of the structures of MHC molecules and the complexes of MHC/TCR with specificity for many different peptides. In recent years, the structures of MHC-I molecules complexed with chaperones that assist in peptide loading have been revealed by X-ray crystallography and cryogenic electron microscopy. These structures have been further studied using mutagenesis, molecular dynamics and NMR approaches. This review summarizes the current structures and dynamic principles that govern peptide exchange as these relate to the process of antigen presentation.
Keywords: MHC; MHC-I/TAPBPR; MHC-I/tapasin; PLC; antigen presentation; chaperones; major histocompatibility complex; peptide exchange; structural immunology.
open access.
Figures







Similar articles
-
CryoEM structure of an MHC-I/TAPBPR peptide-bound intermediate reveals the mechanism of antigen proofreading.Proc Natl Acad Sci U S A. 2025 Jan 14;122(2):e2416992122. doi: 10.1073/pnas.2416992122. Epub 2025 Jan 9. Proc Natl Acad Sci U S A. 2025. PMID: 39786927 Free PMC article.
-
Structure and Function of Molecular Chaperones that Govern Immune Peptide Loading.Subcell Biochem. 2019;93:321-337. doi: 10.1007/978-3-030-28151-9_10. Subcell Biochem. 2019. PMID: 31939156 Review.
-
Chaperone function in antigen presentation by MHC class I molecules-tapasin in the PLC and TAPBPR beyond.Front Immunol. 2023 Jun 15;14:1179846. doi: 10.3389/fimmu.2023.1179846. eCollection 2023. Front Immunol. 2023. PMID: 37398669 Free PMC article. Review.
-
Structural aspects of chaperone-mediated peptide loading in the MHC-I antigen presentation pathway.Crit Rev Biochem Mol Biol. 2019 Apr;54(2):164-173. doi: 10.1080/10409238.2019.1610352. Epub 2019 May 14. Crit Rev Biochem Mol Biol. 2019. PMID: 31084439 Free PMC article. Review.
-
MHC I chaperone complexes shaping immunity.Curr Opin Immunol. 2019 Jun;58:9-15. doi: 10.1016/j.coi.2019.01.001. Epub 2019 Feb 14. Curr Opin Immunol. 2019. PMID: 30771631 Review.
Cited by
-
CryoEM structure of an MHC-I/TAPBPR peptide-bound intermediate reveals the mechanism of antigen proofreading.Proc Natl Acad Sci U S A. 2025 Jan 14;122(2):e2416992122. doi: 10.1073/pnas.2416992122. Epub 2025 Jan 9. Proc Natl Acad Sci U S A. 2025. PMID: 39786927 Free PMC article.
-
The potential applications of peptide-loading complex in cancer treatment.Front Immunol. 2025 Mar 3;16:1526137. doi: 10.3389/fimmu.2025.1526137. eCollection 2025. Front Immunol. 2025. PMID: 40098955 Free PMC article. Review.
References
-
- Berman, H. M., Battistuz, T., Bhat, T. N., Bluhm, W. F., Bourne, P. E., Burkhardt, K., Feng, Z., Gilliland, G. L., Iype, L., Jain, S., Fagan, P., Marvin, J., Padilla, D., Ravichandran, V., Schneider, B., Thanki, N., Weissig, H., Westbrook, J. D. & Zardecki, C. (2002). Acta Cryst. D58, 899–907. - PubMed
-
- Blees, A., Januliene, D., Hofmann, T., Koller, N., Schmidt, C., Trowitzsch, S., Moeller, A. & Tampé, R. (2017). Nature, 551, 525–528. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

