Structural insights into the molecular mechanism of phytoplasma immunodominant membrane protein
- PMID: 38656311
- PMCID: PMC11067747
- DOI: 10.1107/S2052252524003075
Structural insights into the molecular mechanism of phytoplasma immunodominant membrane protein
Abstract
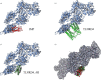
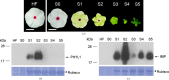
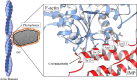
Immunodominant membrane protein (IMP) is a prevalent membrane protein in phytoplasma and has been confirmed to be an F-actin-binding protein. However, the intricate molecular mechanisms that govern the function of IMP require further elucidation. In this study, the X-ray crystallographic structure of IMP was determined and insights into its interaction with plant actin are provided. A comparative analysis with other proteins demonstrates that IMP shares structural homology with talin rod domain-containing protein 1 (TLNRD1), which also functions as an F-actin-binding protein. Subsequent molecular-docking studies of IMP and F-actin reveal that they possess complementary surfaces, suggesting a stable interaction. The low potential energy and high confidence score of the IMP-F-actin binding model indicate stable binding. Additionally, by employing immunoprecipitation and mass spectrometry, it was discovered that IMP serves as an interaction partner for the phytoplasmal effector causing phyllody 1 (PHYL1). It was then shown that both IMP and PHYL1 are highly expressed in the S2 stage of peanut witches' broom phytoplasma-infected Catharanthus roseus. The association between IMP and PHYL1 is substantiated through in vivo immunoprecipitation, an in vitro cross-linking assay and molecular-docking analysis. Collectively, these findings expand the current understanding of IMP interactions and enhance the comprehension of the interaction of IMP with plant F-actin. They also unveil a novel interaction pathway that may influence phytoplasma pathogenicity and host plant responses related to PHYL1. This discovery could pave the way for the development of new strategies to overcome phytoplasma-related plant diseases.
Keywords: X-ray crystallography; actin-binding proteins; immunodominant membrane proteins; phytoplasma; protein structure; α-helix bundles.
open access.
Figures




References
-
- Bonnot, F., de Franqueville, H. & Lourenço, E. (2010). Phytopathology, 100, 300–312. - PubMed
-
- Boonrod, K., Munteanu, B., Jarausch, B., Jarausch, W. & Krczal, G. (2012). Mol. Plant Microbe Interact. 25, 889–895. - PubMed
-
- Constable, F. E. (2009). Phytoplasmas: Genomes, Plant Hosts and Vectors, edited by P. G. Weintraub & P. Jones, pp. 188–212. Wallingford: CABI.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

