Explainable AI for CHO cell culture media optimization and prediction of critical quality attribute
- PMID: 38656382
- PMCID: PMC11043154
- DOI: 10.1007/s00253-024-13147-w
Explainable AI for CHO cell culture media optimization and prediction of critical quality attribute
Abstract
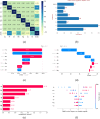
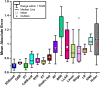
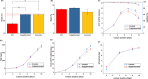
Cell culture media play a critical role in cell growth and propagation by providing a substrate; media components can also modulate the critical quality attributes (CQAs). However, the inherent complexity of the cell culture media makes unraveling the impact of the various media components on cell growth and CQAs non-trivial. In this study, we demonstrate an end-to-end machine learning framework for media component selection and prediction of CQAs. The preliminary dataset for feature selection was generated by performing CHO-GS (-/-) cell culture in media formulations with varying metal ion concentrations. Acidic and basic charge variant composition of the innovator product (24.97 ± 0.54% acidic and 11.41 ± 1.44% basic) was chosen as the target variable to evaluate the media formulations. Pearson's correlation coefficient and random forest-based techniques were used for feature ranking and feature selection for the prediction of acidic and basic charge variants. Furthermore, a global interpretation analysis using SHapley Additive exPlanations was utilized to select optimal features by evaluating the contributions of each feature in the extracted vectors. Finally, the medium combinations were predicted by employing fifteen different regression models and utilizing a grid search and random search cross-validation for hyperparameter optimization. Experimental results demonstrate that Fe and Zn significantly impact the charge variant profile. This study aims to offer insights that are pertinent to both innovators seeking to establish a complete pipeline for media development and optimization and biosimilar-based manufacturers who strive to demonstrate the analytical and functional biosimilarity of their products to the innovator. KEY POINTS: • Developed a framework for optimizing media components and prediction of CQA. • SHAP enhances global interpretability, aiding informed decision-making. • Fifteen regression models were employed to predict medium combinations.
Keywords: Biosimilar; Charge variants; Feature ranking; Feature selection; Machine learning; Media development.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
QbD-guided pharmaceutical development of Pembrolizumab biosimilar candidate PSG-024 propelled to industry meeting primary requirements of comparability to Keytruda®.Eur J Pharm Sci. 2022 Jun 1;173:106171. doi: 10.1016/j.ejps.2022.106171. Epub 2022 Apr 1. Eur J Pharm Sci. 2022. PMID: 35378209
-
Improving titer while maintaining quality of final formulated drug substance via optimization of CHO cell culture conditions in low-iron chemically defined media.MAbs. 2018 Apr;10(3):488-499. doi: 10.1080/19420862.2018.1433978. Epub 2018 Feb 20. MAbs. 2018. PMID: 29388872 Free PMC article.
-
Use of spectroscopic process analytical technology for rapid quality evaluation during preparation of CHO cell culture media.Biotechnol Prog. 2024 Sep-Oct;40(5):e3477. doi: 10.1002/btpr.3477. Epub 2024 May 3. Biotechnol Prog. 2024. PMID: 38699906
-
Challenges in developing cell culture media using machine learning.Biotechnol Adv. 2024 Jan-Feb;70:108293. doi: 10.1016/j.biotechadv.2023.108293. Epub 2023 Nov 19. Biotechnol Adv. 2024. PMID: 37984683 Review.
-
Cell culture media for recombinant protein expression in Chinese hamster ovary (CHO) cells: History, key components, and optimization strategies.Biotechnol Prog. 2018 Nov;34(6):1407-1426. doi: 10.1002/btpr.2706. Epub 2018 Oct 5. Biotechnol Prog. 2018. PMID: 30290072 Review.
Cited by
-
Wheels turning: CHO cell modeling moves into a digital biomanufacturing era: Subtitle: CHO Metabolic Modeling.Comput Struct Biotechnol J. 2025 Jun 23;27:2796-2813. doi: 10.1016/j.csbj.2025.06.035. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40677243 Free PMC article. Review.
-
Machine learning-driven optimization of culture conditions and media components to mitigate charge heterogeneity in monoclonal antibody production: current advances and future perspectives.MAbs. 2025 Dec;17(1):2547084. doi: 10.1080/19420862.2025.2547084. Epub 2025 Aug 14. MAbs. 2025. PMID: 40810344 Free PMC article. Review.
-
Characterization and design of dipeptide media formulation for scalable therapeutic production.Appl Microbiol Biotechnol. 2025 Jan 14;109(1):7. doi: 10.1007/s00253-024-13402-0. Appl Microbiol Biotechnol. 2025. PMID: 39808320 Free PMC article. Review.
-
Innovative Formulation Strategies for Biosimilars: Trends Focused on Buffer-Free Systems, Safety, Regulatory Alignment, and Intellectual Property Challenges.Pharmaceuticals (Basel). 2025 Jun 17;18(6):908. doi: 10.3390/ph18060908. Pharmaceuticals (Basel). 2025. PMID: 40573303 Free PMC article. Review.
-
Recent advances in culture medium design for enhanced production of monoclonal antibodies in CHO cells: A comparative study of machine learning and systems biology approaches.Biotechnol Adv. 2025 Jan-Feb;78:108480. doi: 10.1016/j.biotechadv.2024.108480. Epub 2024 Nov 19. Biotechnol Adv. 2025. PMID: 39571767 Review.
References
-
- Altmann A, Toloşi L, Sander O, Lengauer T (2010) Permutation importance: a corrected feature importance measure. Bioinformatics 26:1340–1347. 10.1093/BIOINFORMATICS/BTQ134 - PubMed
-
- Basu V (2020) Prediction of stellar age with the help of extra-trees regressor in machine learning. Proceedings of the International Conference on Innovative Computing and Communications (ICICC) 2020, Available at SSRN: https://ssrn.com/abstract=3563397 or 10.2139/ssrn.3563397
-
- Breiman L (2001) Random forests. Mach Learn 45:5–32. 10.1023/A:1010933404324
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

