Interplay of condensation and chromatin binding underlies BRD4 targeting
- PMID: 38656803
- PMCID: PMC11238092
- DOI: 10.1091/mbc.E24-01-0046
Interplay of condensation and chromatin binding underlies BRD4 targeting
Abstract
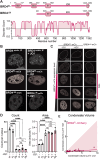
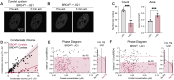
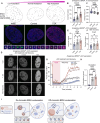
Nuclear compartments form via biomolecular phase separation, mediated through multivalent properties of biomolecules concentrated within condensates. Certain compartments are associated with specific chromatin regions, including transcriptional initiation condensates, which are composed of transcription factors and transcriptional machinery, and form at acetylated regions including enhancer and promoter loci. While protein self-interactions, especially within low-complexity and intrinsically disordered regions, are known to mediate condensation, the role of substrate-binding interactions in regulating the formation and function of biomolecular condensates is underexplored. Here, utilizing live-cell experiments in parallel with coarse-grained simulations, we investigate how chromatin interaction of the transcriptional activator BRD4 modulates its condensate formation. We find that both kinetic and thermodynamic properties of BRD4 condensation are affected by chromatin binding: nucleation rate is sensitive to BRD4-chromatin interactions, providing an explanation for the selective formation of BRD4 condensates at acetylated chromatin regions, and thermodynamically, multivalent acetylated chromatin sites provide a platform for BRD4 clustering below the concentration required for off-chromatin condensation. This provides a molecular and physical explanation of the relationship between nuclear condensates and epigenetically modified chromatin that results in their mutual spatiotemporal regulation, suggesting that epigenetic modulation is an important mechanism by which the cell targets transcriptional condensates to specific chromatin loci.
Figures



Similar articles
-
Organization of Chromatin by Intrinsic and Regulated Phase Separation.Cell. 2019 Oct 3;179(2):470-484.e21. doi: 10.1016/j.cell.2019.08.037. Epub 2019 Sep 19. Cell. 2019. PMID: 31543265 Free PMC article.
-
RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation.Nat Struct Mol Biol. 2018 Aug;25(8):687-697. doi: 10.1038/s41594-018-0102-0. Epub 2018 Aug 3. Nat Struct Mol Biol. 2018. PMID: 30076409 Free PMC article.
-
Signal-induced Brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation.Nucleic Acids Res. 2011 Dec;39(22):9592-604. doi: 10.1093/nar/gkr698. Epub 2011 Sep 2. Nucleic Acids Res. 2011. PMID: 21890894 Free PMC article.
-
The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation.J Biol Chem. 2007 May 4;282(18):13141-5. doi: 10.1074/jbc.R700001200. Epub 2007 Feb 28. J Biol Chem. 2007. PMID: 17329240 Review.
-
Therapeutic targeting of BET bromodomain and other epigenetic acetylrecognition domain-containing factors.Curr Opin Genet Dev. 2024 Jun;86:102181. doi: 10.1016/j.gde.2024.102181. Epub 2024 Apr 2. Curr Opin Genet Dev. 2024. PMID: 38564841 Review.
Cited by
-
BRD4 binds the nucleosome via both histone and DNA interactions.bioRxiv [Preprint]. 2025 May 30:2025.05.29.656846. doi: 10.1101/2025.05.29.656846. bioRxiv. 2025. PMID: 40501777 Free PMC article. Preprint.
-
Interfacial effects determine nonequilibrium phase behaviors in chemically driven fluids.Proc Natl Acad Sci U S A. 2025 Jul 29;122(30):e2501145122. doi: 10.1073/pnas.2501145122. Epub 2025 Jul 23. Proc Natl Acad Sci U S A. 2025. PMID: 40699931
-
RNA polymerase II transcription compartments - from factories to condensates.Nat Rev Genet. 2025 Jun 19. doi: 10.1038/s41576-025-00859-6. Online ahead of print. Nat Rev Genet. 2025. PMID: 40537661 Review.
-
Control of Inflammatory Response by Tissue Microenvironment.bioRxiv [Preprint]. 2025 May 30:2024.05.10.592432. doi: 10.1101/2024.05.10.592432. bioRxiv. 2025. PMID: 38798655 Free PMC article. Preprint.
-
A Minireview on BET Inhibitors: Beyond Bromodomain Targeting.Biomedicines. 2025 Mar 1;13(3):594. doi: 10.3390/biomedicines13030594. Biomedicines. 2025. PMID: 40149571 Free PMC article. Review.
References
-
- Alekseyenko AA, Walsh EM, Zee BM, Pakozdi T, Hsi P, Lemieux ME, Cin PD, Ince TA, Kharchenko PV, Kuroda M, French CA (2017). Ectopic protein interactions within BRD4-chromatin complexes drive oncogenic megadomain formation in NUT midline carcinoma. Proc Natl Acad Sci USA 114, E4184–4192. - PMC - PubMed
-
- Bianchi E, Largo J, Tartaglia P, Zaccarelli E, Sciortino F (2006). Phase diagram of patchy colloids: towards empty liquids. Phys Rev Lett 97, 168301. - PubMed
-
- Boisvert F-M, van Koningsbruggen S, Navascués J, Lamond AI (2007). The multifunctional nucleolus. Nat Rev Mol Cell Biol 8, 1021–574– 585. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

