Polycomb protein binding and looping in the ON transcriptional state
- PMID: 38657072
- PMCID: PMC11042752
- DOI: 10.1126/sciadv.adn1837
Polycomb protein binding and looping in the ON transcriptional state
Abstract
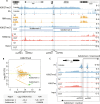
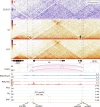
Polycomb group (PcG) proteins mediate epigenetic silencing of important developmental genes by modifying histones and compacting chromatin through two major protein complexes, PRC1 and PRC2. These complexes are recruited to DNA by CpG islands (CGIs) in mammals and Polycomb response elements (PREs) in Drosophila. When PcG target genes are turned OFF, PcG proteins bind to PREs or CGIs, and PREs serve as anchors that loop together and stabilize gene silencing. Here, we address which PcG proteins bind to PREs and whether PREs mediate looping when their targets are in the ON transcriptional state. While the binding of most PcG proteins decreases at PREs in the ON state, one PRC1 component, Ph, remains bound. Further, PREs can loop to each other and with presumptive enhancers in the ON state and, like CGIs, may act as tethering elements between promoters and enhancers. Overall, our data suggest that PREs are important looping elements for developmental loci in both the ON and OFF states.
Figures



Update of
-
Polycomb protein binding and looping mediated by Polycomb Response Elements in the ON transcriptional state.bioRxiv [Preprint]. 2023 Nov 4:2023.11.02.565256. doi: 10.1101/2023.11.02.565256. bioRxiv. 2023. Update in: Sci Adv. 2024 Apr 26;10(17):eadn1837. doi: 10.1126/sciadv.adn1837. PMID: 38076900 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

