Autonomous circadian rhythms in the human hepatocyte regulate hepatic drug metabolism and inflammatory responses
- PMID: 38657074
- PMCID: PMC11042741
- DOI: 10.1126/sciadv.adm9281
Autonomous circadian rhythms in the human hepatocyte regulate hepatic drug metabolism and inflammatory responses
Abstract
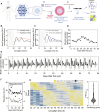
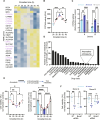
Critical aspects of physiology and cell function exhibit self-sustained ~24-hour variations termed circadian rhythms. In the liver, circadian rhythms play fundamental roles in maintaining organ homeostasis. Here, we established and characterized an in vitro liver experimental system in which primary human hepatocytes display self-sustained oscillations. By generating gene expression profiles of these hepatocytes over time, we demonstrated that their transcriptional state is dynamic across 24 hours and identified a set of cycling genes with functions related to inflammation, drug metabolism, and energy homeostasis. We designed and tested a treatment protocol to minimize atorvastatin- and acetaminophen-induced hepatotoxicity. Last, we documented circadian-dependent induction of pro-inflammatory cytokines when triggered by LPS, IFN-β, or Plasmodium infection in human hepatocytes. Collectively, our findings emphasize that the phase of the circadian cycle has a robust impact on the efficacy and toxicity of drugs, and we provide a test bed to study the timing and magnitude of inflammatory responses over the course of infection in human liver.
Figures



References
-
- Skarke C., Lahens N. F., Rhoades S. D., Campbell A., Bittinger K., Bailey A., Hoffmann C., Olson R. S., Chen L., Yang G., Price T. S., Moore J. H., Bushman F. D., Greene C. S., Grant G. R., Weljie A. M., Fitzgerald G. A., A pilot characterization of the human chronobiome. Sci. Rep. 7, 17141 (2017). - PMC - PubMed
-
- Cederroth C. R., Albrecht U., Bass J., Brown S. A., Dyhrfjeld-Johnsen J., Gachon F., Green C. B., Hastings M. H., Helfrich-Förster C., Hogenesch J. B., Lévi F., Loudon A., Lundkvist G. B., Meijer J. H., Rosbash M., Takahashi J. S., Young M., Canlon B., Medicine in the fourth dimension. Cell Metab. 30, 238–250 (2019). - PMC - PubMed
-
- Qian D. C., Kleber T., Brammer B., Xu K. M., Switchenko J. M., Janopaul-Naylor J. R., Zhong J., Yushak M. L., Harvey R. D., Paulos C. M., Lawson D. H., Khan M. K., Kudchadkar R. R., Buchwald Z. S., Effect of immunotherapy time-of-day infusion on overall survival among patients with advanced melanoma in the USA (MEMOIR): A propensity score-matched analysis of a single-centre, longitudinal study. Lancet Oncol. 22, 1777–1786 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

