Vacuolar degradation of plant organelles
- PMID: 38657116
- PMCID: PMC11371181
- DOI: 10.1093/plcell/koae128
Vacuolar degradation of plant organelles
Abstract
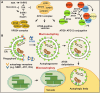
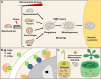
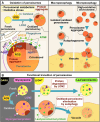
Plants continuously remodel and degrade their organelles due to damage from their metabolic activities and environmental stressors, as well as an integral part of their cell differentiation programs. Whereas certain organelles use local hydrolytic enzymes for limited remodeling, most of the pathways that control the partial or complete dismantling of organelles rely on vacuolar degradation. Specifically, selective autophagic pathways play a crucial role in recognizing and sorting plant organelle cargo for vacuolar clearance, especially under cellular stress conditions induced by factors like heat, drought, and damaging light. In these short reviews, we discuss the mechanisms that control the vacuolar degradation of chloroplasts, mitochondria, endoplasmic reticulum, Golgi, and peroxisomes, with an emphasis on autophagy, recently discovered selective autophagy receptors for plant organelles, and crosstalk with other catabolic pathways.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

