Enhancing Standard of Care Chemotherapy Efficacy Using DNA-Dependent Protein Kinase (DNA-PK) Inhibition in Preclinical Models of Ewing Sarcoma
- PMID: 38657228
- PMCID: PMC11293986
- DOI: 10.1158/1535-7163.MCT-23-0641
Enhancing Standard of Care Chemotherapy Efficacy Using DNA-Dependent Protein Kinase (DNA-PK) Inhibition in Preclinical Models of Ewing Sarcoma
Abstract
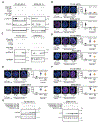
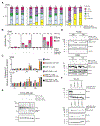
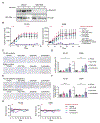
Disruption of DNA damage repair via impaired homologous recombination is characteristic of Ewing sarcoma (EWS) cells. We hypothesize that this disruption results in increased reliance on nonhomologous end joining to repair DNA damage. In this study, we investigated if pharmacologic inhibition of the enzyme responsible for nonhomologous end joining, the DNA-PK holoenzyme, alters the response of EWS cells to genotoxic standard of care chemotherapy. We used analyses of cell viability and proliferation to investigate the effects of clinical DNA-PK inhibitors (DNA-PKi) in combination with six therapeutic or experimental agents for EWS. We performed calculations of synergy using the Loewe additivity model. Immunoblotting evaluated treatment effects on DNA-PK, DNA damage, and apoptosis. Flow cytometric analyses evaluated effects on cell cycle and fate. We used orthotopic xenograft models to interrogate tolerability, drug mechanism, and efficacy in vivo. DNA-PKi demonstrated on-target activity, reducing phosphorylated DNA-PK levels in EWS cells. DNA-PKi sensitized EWS cell lines to agents that function as topoisomerase 2 (TOP2) poisons and enhanced the DNA damage induced by TOP2 poisons. Nanomolar concentrations of single-agent TOP2 poisons induced G2M arrest and little apoptotic response while adding DNA-PKi-mediated apoptosis. In vivo, the combination of AZD7648 and etoposide had limited tolerability but resulted in enhanced DNA damage, apoptosis, and EWS tumor shrinkage. The combination of DNA-PKi with standard of care TOP2 poisons in EWS models is synergistic, enhances DNA damage and cell death, and may form the basis of a promising future therapeutic strategy for EWS.
©2024 American Association for Cancer Research.
Conflict of interest statement
Figures


Similar articles
-
Targeted inhibition of histone deacetylase leads to suppression of Ewing sarcoma tumor growth through an unappreciated EWS-FLI1/HDAC3/HSP90 signaling axis.J Mol Med (Berl). 2019 Jul;97(7):957-972. doi: 10.1007/s00109-019-01782-0. Epub 2019 Apr 25. J Mol Med (Berl). 2019. PMID: 31025088 Free PMC article.
-
TAE226, a dual inhibitor of focal adhesion kinase and insulin-like growth factor-I receptor, is effective for Ewing sarcoma.Cancer Med. 2019 Dec;8(18):7809-7821. doi: 10.1002/cam4.2647. Epub 2019 Nov 6. Cancer Med. 2019. PMID: 31692287 Free PMC article.
-
Cell Context Is the Third Axis of Synergy for the Combination of ATR Inhibition and Cisplatin in Ewing Sarcoma.Clin Cancer Res. 2024 Aug 15;30(16):3533-3548. doi: 10.1158/1078-0432.CCR-23-3063. Clin Cancer Res. 2024. PMID: 38506712
-
Targeting the EWS-ETS transcriptional program by BET bromodomain inhibition in Ewing sarcoma.Oncotarget. 2016 Jan 12;7(2):1451-63. doi: 10.18632/oncotarget.6385. Oncotarget. 2016. PMID: 26623725 Free PMC article.
-
Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441.Cancer Res. 2006 May 15;66(10):5354-62. doi: 10.1158/0008-5472.CAN-05-4275. Cancer Res. 2006. PMID: 16707462
Cited by
-
Therapeutic Targeting of DNA Repair Pathways in Pediatric Extracranial Solid Tumors: Current State and Implications for Immunotherapy.Cancers (Basel). 2024 Apr 25;16(9):1648. doi: 10.3390/cancers16091648. Cancers (Basel). 2024. PMID: 38730598 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

