C9ORF72 Deficiency Results in Neurodegeneration in the Zebrafish Retina
- PMID: 38658168
- PMCID: PMC11209673
- DOI: 10.1523/JNEUROSCI.2128-23.2024
C9ORF72 Deficiency Results in Neurodegeneration in the Zebrafish Retina
Abstract
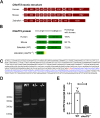
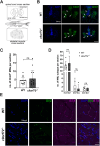
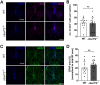
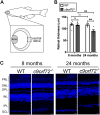
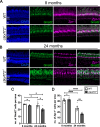
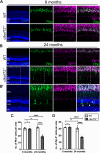
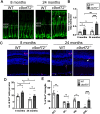
Hexanucleotide repeat expansions within the gene C9ORF72 are the most common cause of the neurodegenerative diseases amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). This disease-causing expansion leads to a reduction in C9ORF72 expression levels in patients, suggesting loss of C9ORF72 function could contribute to disease. To further understand the consequences of C9ORF72 deficiency in vivo, we generated a c9orf72 mutant zebrafish line. Analysis of the adult female spinal cords revealed no appreciable neurodegenerative pathology such as loss of motor neurons or increased levels of neuroinflammation. However, detailed examination of adult female c9orf72-/- retinas showed prominent neurodegenerative features, including a decrease in retinal thickness, gliosis, and an overall reduction in neurons of all subtypes. Analysis of rod and cone cells within the photoreceptor layer showed a disturbance in their outer segment structure and rhodopsin mislocalization from rod outer segments to their cell bodies and synaptic terminals. Thus, C9ORF72 may play a previously unappreciated role in retinal homeostasis and suggests C9ORF72 deficiency can induce tissue specific neuronal loss.
Keywords: ALS; C9ORF72; FTD; neurodegeneration; zebrafish.
Copyright © 2024 Jaroszynska et al.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
