Flexible TAM requirement of TnpB enables efficient single-nucleotide editing with expanded targeting scope
- PMID: 38658536
- PMCID: PMC11043419
- DOI: 10.1038/s41467-024-47697-4
Flexible TAM requirement of TnpB enables efficient single-nucleotide editing with expanded targeting scope
Abstract
TnpBs encoded by the IS200/IS605 family transposon are among the most abundant prokaryotic proteins from which type V CRISPR-Cas nucleases may have evolved. Since bacterial TnpBs can be programmed for RNA-guided dsDNA cleavage in the presence of a transposon-adjacent motif (TAM), these nucleases hold immense promise for genome editing. However, the activity and targeting specificity of TnpB in homology-directed gene editing remain unknown. Here we report that a thermophilic archaeal TnpB enables efficient gene editing in the natural host. Interestingly, the TnpB has different TAM requirements for eliciting cell death and for facilitating gene editing. By systematically characterizing TAM variants, we reveal that the TnpB recognizes a broad range of TAM sequences for gene editing including those that do not elicit apparent cell death. Importantly, TnpB shows a very high targeting specificity on targets flanked by a weak TAM. Taking advantage of this feature, we successfully leverage TnpB for efficient single-nucleotide editing with templated repair. The use of different weak TAM sequences not only facilitates more flexible gene editing with increased cell survival, but also greatly expands targeting scopes, and this strategy is probably applicable to diverse CRISPR-Cas systems.
© 2024. The Author(s).
Conflict of interest statement
X.F., Q.S., R.X., J.L. and P.Z. are co-inventors on a patent application (CN202310186099.8) filled by Shandong University relating to this work. All other authors declare no competing interests.
Figures
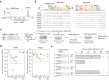
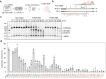




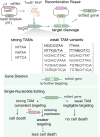
Similar articles
-
Evolutionary mining and functional characterization of TnpB nucleases identify efficient miniature genome editors.Nat Biotechnol. 2024 May;42(5):745-757. doi: 10.1038/s41587-023-01857-x. Epub 2023 Jun 29. Nat Biotechnol. 2024. PMID: 37386294
-
Experimental strategy for characterization of novel TnpB orthologs.Methods Enzymol. 2025;712:183-195. doi: 10.1016/bs.mie.2025.01.056. Epub 2025 Mar 5. Methods Enzymol. 2025. PMID: 40121072
-
Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease.Nature. 2021 Nov;599(7886):692-696. doi: 10.1038/s41586-021-04058-1. Epub 2021 Oct 7. Nature. 2021. PMID: 34619744 Free PMC article.
-
Genome editing using CRISPR, CAST, and Fanzor systems.Mol Cells. 2024 Jul;47(7):100086. doi: 10.1016/j.mocell.2024.100086. Epub 2024 Jun 21. Mol Cells. 2024. PMID: 38909984 Free PMC article. Review.
-
Biology and applications of CRISPR-Cas12 and transposon-associated homologs.Nat Biotechnol. 2024 Dec;42(12):1807-1821. doi: 10.1038/s41587-024-02485-9. Epub 2024 Dec 4. Nat Biotechnol. 2024. PMID: 39633151 Review.
Cited by
-
Characterization of the genome editing with miniature DNA nucleases TnpB and IscB in Escherichia coli strains.Commun Biol. 2025 Feb 19;8(1):261. doi: 10.1038/s42003-025-07521-1. Commun Biol. 2025. PMID: 39972101 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- 32001022/National Natural Science Foundation of China (National Science Foundation of China)
- 32270040/National Natural Science Foundation of China (National Science Foundation of China)
- 2020YFA0906800/Ministry of Science and Technology of the People's Republic of China (Chinese Ministry of Science and Technology)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

