Single cell tracing of Pomc neurons reveals recruitment of 'Ghost' subtypes with atypical identity in a mouse model of obesity
- PMID: 38658557
- PMCID: PMC11043070
- DOI: 10.1038/s41467-024-47877-2
Single cell tracing of Pomc neurons reveals recruitment of 'Ghost' subtypes with atypical identity in a mouse model of obesity
Abstract
The hypothalamus contains a remarkable diversity of neurons that orchestrate behavioural and metabolic outputs in a highly plastic manner. Neuronal diversity is key to enabling hypothalamic functions and, according to the neuroscience dogma, it is predetermined during embryonic life. Here, by combining lineage tracing of hypothalamic pro-opiomelanocortin (Pomc) neurons with single-cell profiling approaches in adult male mice, we uncovered subpopulations of 'Ghost' neurons endowed with atypical molecular and functional identity. Compared to 'classical' Pomc neurons, Ghost neurons exhibit negligible Pomc expression and are 'invisible' to available neuroanatomical approaches and promoter-based reporter mice for studying Pomc biology. Ghost neuron numbers augment in diet-induced obese mice, independent of neurogenesis or cell death, but weight loss can reverse this shift. Our work challenges the notion of fixed, developmentally programmed neuronal identities in the mature hypothalamus and highlight the ability of specialised neurons to reversibly adapt their functional identity to adult-onset obesogenic stimuli.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
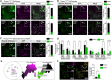
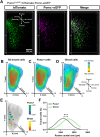

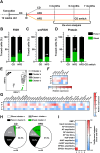
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

