Functional synergy of a human-specific and an ape-specific metabolic regulator in human neocortex development
- PMID: 38658571
- PMCID: PMC11043075
- DOI: 10.1038/s41467-024-47437-8
Functional synergy of a human-specific and an ape-specific metabolic regulator in human neocortex development
Abstract
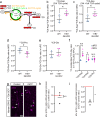
Metabolism has recently emerged as a major target of genes implicated in the evolutionary expansion of human neocortex. One such gene is the human-specific gene ARHGAP11B. During human neocortex development, ARHGAP11B increases the abundance of basal radial glia, key progenitors for neocortex expansion, by stimulating glutaminolysis (glutamine-to-glutamate-to-alpha-ketoglutarate) in mitochondria. Here we show that the ape-specific protein GLUD2 (glutamate dehydrogenase 2), which also operates in mitochondria and converts glutamate-to-αKG, enhances ARHGAP11B's ability to increase basal radial glia abundance. ARHGAP11B + GLUD2 double-transgenic bRG show increased production of aspartate, a metabolite essential for cell proliferation, from glutamate via alpha-ketoglutarate and the TCA cycle. Hence, during human evolution, a human-specific gene exploited the existence of another gene that emerged during ape evolution, to increase, via concerted changes in metabolism, progenitor abundance and neocortex size.
© 2024. The Author(s).
Conflict of interest statement
O.H. occasionally serves on advisory boards for Bayer AG and Gedeon Richter and has designed and lectured at educational events for these companies. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

