Skeletal muscle differentiation induces wide-ranging nucleosome repositioning in muscle gene promoters
- PMID: 38658615
- PMCID: PMC11043329
- DOI: 10.1038/s41598-024-60236-x
Skeletal muscle differentiation induces wide-ranging nucleosome repositioning in muscle gene promoters
Abstract
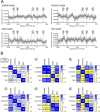
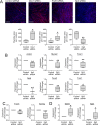
In a previous report, we demonstrated that Cbx1, PurB and Sp3 inhibited cardiac muscle differentiation by increasing nucleosome density around cardiac muscle gene promoters. Since cardiac and skeletal muscle express many of the same proteins, we asked if Cbx1, PurB and Sp3 similarly regulated skeletal muscle differentiation. In a C2C12 model of skeletal muscle differentiation, Cbx1 and PurB knockdown increased myotube formation. In contrast, Sp3 knockdown inhibited myotube formation, suggesting that Sp3 played opposing roles in cardiac muscle and skeletal muscle differentiation. Consistent with this finding, Sp3 knockdown also inhibited various muscle-specific genes. The Cbx1, PurB and Sp3 proteins are believed to influence gene-expression in part by altering nucleosome position. Importantly, we developed a statistical approach to determine if changes in nucleosome positioning were significant and applied it to understanding the architecture of muscle-specific genes. Through this novel statistical approach, we found that during myogenic differentiation, skeletal muscle-specific genes undergo a set of unique nucleosome changes which differ significantly from those shown in commonly expressed muscle genes. While Sp3 binding was associated with nucleosome loss, there appeared no correlation with the aforementioned nucleosome changes. In summary, we have identified a novel role for Sp3 in skeletal muscle differentiation and through the application of quantifiable MNase-seq have discovered unique fingerprints of nucleosome changes for various classes of muscle genes during myogenic differentiation.
Keywords: Cardiac muscle; ChIP-seq; MNase-seq; Skeletal muscle; Sp3.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

