Antisense oligonucleotide therapeutic approach for Timothy syndrome
- PMID: 38658687
- PMCID: PMC11043036
- DOI: 10.1038/s41586-024-07310-6
Antisense oligonucleotide therapeutic approach for Timothy syndrome
Abstract
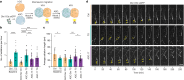
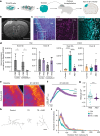
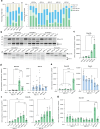
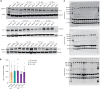
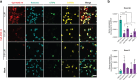
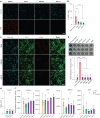
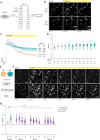
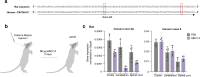
Timothy syndrome (TS) is a severe, multisystem disorder characterized by autism, epilepsy, long-QT syndrome and other neuropsychiatric conditions1. TS type 1 (TS1) is caused by a gain-of-function variant in the alternatively spliced and developmentally enriched CACNA1C exon 8A, as opposed to its counterpart exon 8. We previously uncovered several phenotypes in neurons derived from patients with TS1, including delayed channel inactivation, prolonged depolarization-induced calcium rise, impaired interneuron migration, activity-dependent dendrite retraction and an unanticipated persistent expression of exon 8A2-6. We reasoned that switching CACNA1C exon utilization from 8A to 8 would represent a potential therapeutic strategy. Here we developed antisense oligonucleotides (ASOs) to effectively decrease the inclusion of exon 8A in human cells both in vitro and, following transplantation, in vivo. We discovered that the ASO-mediated switch from exon 8A to 8 robustly rescued defects in patient-derived cortical organoids and migration in forebrain assembloids. Leveraging a transplantation platform previously developed7, we found that a single intrathecal ASO administration rescued calcium changes and in vivo dendrite retraction of patient neurons, suggesting that suppression of CACNA1C exon 8A expression is a potential treatment for TS1. Broadly, these experiments illustrate how a multilevel, in vivo and in vitro stem cell model-based approach can identify strategies to reverse disease-relevant neural pathophysiology.
© 2024. The Author(s).
Conflict of interest statement
Stanford University holds patents for the generation of cortical organoids/spheroids and assembloids (listing S.P.P., F.B. as inventors), a patent application for ASO (listing S.P.P., X.C. and F.B. as inventors) and a patent application for transplantation of organoids (listing S.P.P. and O.R. as inventors).
Figures







Comment in
-
ASO to treat Timothy syndrome.Nat Rev Drug Discov. 2024 Jun;23(6):420. doi: 10.1038/d41573-024-00075-7. Nat Rev Drug Discov. 2024. PMID: 38714847 No abstract available.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

