PGE2 limits effector expansion of tumour-infiltrating stem-like CD8+ T cells
- PMID: 38658748
- PMCID: PMC11078747
- DOI: 10.1038/s41586-024-07254-x
PGE2 limits effector expansion of tumour-infiltrating stem-like CD8+ T cells
Abstract
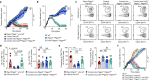
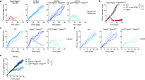
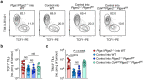
Cancer-specific TCF1+ stem-like CD8+ T cells can drive protective anticancer immunity through expansion and effector cell differentiation1-4; however, this response is dysfunctional in tumours. Current cancer immunotherapies2,5-9 can promote anticancer responses through TCF1+ stem-like CD8+ T cells in some but not all patients. This variation points towards currently ill-defined mechanisms that limit TCF1+CD8+ T cell-mediated anticancer immunity. Here we demonstrate that tumour-derived prostaglandin E2 (PGE2) restricts the proliferative expansion and effector differentiation of TCF1+CD8+ T cells within tumours, which promotes cancer immune escape. PGE2 does not affect the priming of TCF1+CD8+ T cells in draining lymph nodes. PGE2 acts through EP2 and EP4 (EP2/EP4) receptor signalling in CD8+ T cells to limit the intratumoural generation of early and late effector T cell populations that originate from TCF1+ tumour-infiltrating CD8+ T lymphocytes (TILs). Ablation of EP2/EP4 signalling in cancer-specific CD8+ T cells rescues their expansion and effector differentiation within tumours and leads to tumour elimination in multiple mouse cancer models. Mechanistically, suppression of the interleukin-2 (IL-2) signalling pathway underlies the PGE2-mediated inhibition of TCF1+ TIL responses. Altogether, we uncover a key mechanism that restricts the IL-2 responsiveness of TCF1+ TILs and prevents anticancer T cell responses that originate from these cells. This study identifies the PGE2-EP2/EP4 axis as a molecular target to restore IL-2 responsiveness in anticancer TILs to achieve cancer immune control.
© 2024. The Author(s).
Conflict of interest statement
J.P.B., S.B.L., A.-M.P., P.K. and S.K. are preparing a patent application that includes work in this manuscript.
Figures










References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

