Emx2 underlies the development and evolution of marsupial gliding membranes
- PMID: 38658750
- PMCID: PMC11062917
- DOI: 10.1038/s41586-024-07305-3
Emx2 underlies the development and evolution of marsupial gliding membranes
Abstract
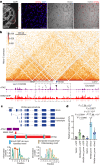
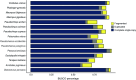
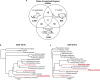
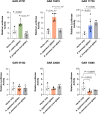
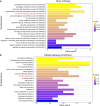
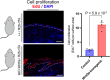
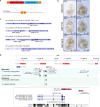
Phenotypic variation among species is a product of evolutionary changes to developmental programs1,2. However, how these changes generate novel morphological traits remains largely unclear. Here we studied the genomic and developmental basis of the mammalian gliding membrane, or patagium-an adaptative trait that has repeatedly evolved in different lineages, including in closely related marsupial species. Through comparative genomic analysis of 15 marsupial genomes, both from gliding and non-gliding species, we find that the Emx2 locus experienced lineage-specific patterns of accelerated cis-regulatory evolution in gliding species. By combining epigenomics, transcriptomics and in-pouch marsupial transgenics, we show that Emx2 is a critical upstream regulator of patagium development. Moreover, we identify different cis-regulatory elements that may be responsible for driving increased Emx2 expression levels in gliding species. Lastly, using mouse functional experiments, we find evidence that Emx2 expression patterns in gliders may have been modified from a pre-existing program found in all mammals. Together, our results suggest that patagia repeatedly originated through a process of convergent genomic evolution, whereby regulation of Emx2 was altered by distinct cis-regulatory elements in independently evolved species. Thus, different regulatory elements targeting the same key developmental gene may constitute an effective strategy by which natural selection has harnessed regulatory evolution in marsupial genomes to generate phenotypic novelty.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Stern DL. Evolutionary developmental biology and the problem of variation. Evolution. 2000;54:1079–1091. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

