Alternative splicing in prostate cancer progression and therapeutic resistance
- PMID: 38658776
- PMCID: PMC11136669
- DOI: 10.1038/s41388-024-03036-x
Alternative splicing in prostate cancer progression and therapeutic resistance
Abstract
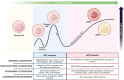
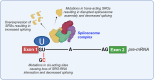
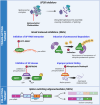
Prostate cancer (CaP) remains the second leading cause of cancer deaths in western men. CaP mortality results from diverse molecular mechanisms that mediate resistance to the standard of care treatments for metastatic disease. Recently, alternative splicing has been recognized as a hallmark of CaP aggressiveness. Alternative splicing events cause treatment resistance and aggressive CaP behavior and are determinants of the emergence of the two major types of late-stage treatment-resistant CaP, namely castration-resistant CaP (CRPC) and neuroendocrine CaP (NEPC). Here, we review recent multi-omics data that are uncovering the complicated landscape of alternative splicing events during CaP progression and the impact that different gene transcript isoforms can have on CaP cell biology and behavior. We discuss renewed insights in the molecular machinery by which alternative splicing occurs and contributes to the failure of systemic CaP therapies. The potential for alternative splicing events to serve as diagnostic markers and/or therapeutic targets is explored. We conclude by considering current challenges and promises associated with splicing-modulating therapies, and their potential for clinical translation into CaP patient care.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, et al. The master neural transcription factor BRN2 Is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017;7:54–71. doi: 10.1158/2159-8290.CD-15-1263. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

