INSaFLU-TELEVIR: an open web-based bioinformatics suite for viral metagenomic detection and routine genomic surveillance
- PMID: 38659008
- PMCID: PMC11044337
- DOI: 10.1186/s13073-024-01334-3
INSaFLU-TELEVIR: an open web-based bioinformatics suite for viral metagenomic detection and routine genomic surveillance
Abstract
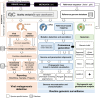
Background: Implementation of clinical metagenomics and pathogen genomic surveillance can be particularly challenging due to the lack of bioinformatics tools and/or expertise. In order to face this challenge, we have previously developed INSaFLU, a free web-based bioinformatics platform for virus next-generation sequencing data analysis. Here, we considerably expanded its genomic surveillance component and developed a new module (TELEVIR) for metagenomic virus identification.
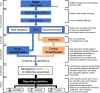
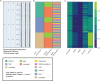
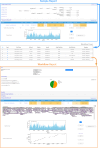
Results: The routine genomic surveillance component was strengthened with new workflows and functionalities, including (i) a reference-based genome assembly pipeline for Oxford Nanopore technologies (ONT) data; (ii) automated SARS-CoV-2 lineage classification; (iii) Nextclade analysis; (iv) Nextstrain phylogeographic and temporal analysis (SARS-CoV-2, human and avian influenza, monkeypox, respiratory syncytial virus (RSV A/B), as well as a "generic" build for other viruses); and (v) algn2pheno for screening mutations of interest. Both INSaFLU pipelines for reference-based consensus generation (Illumina and ONT) were benchmarked against commonly used command line bioinformatics workflows for SARS-CoV-2, and an INSaFLU snakemake version was released. In parallel, a new module (TELEVIR) for virus detection was developed, after extensive benchmarking of state-of-the-art metagenomics software and following up-to-date recommendations and practices in the field. TELEVIR allows running complex workflows, covering several combinations of steps (e.g., with/without viral enrichment or host depletion), classification software (e.g., Kaiju, Kraken2, Centrifuge, FastViromeExplorer), and databases (RefSeq viral genome, Virosaurus, etc.), while culminating in user- and diagnosis-oriented reports. Finally, to potentiate real-time virus detection during ONT runs, we developed findONTime, a tool aimed at reducing costs and the time between sample reception and diagnosis.
Conclusions: The accessibility, versatility, and functionality of INSaFLU-TELEVIR are expected to supply public and animal health laboratories and researchers with a user-oriented and pan-viral bioinformatics framework that promotes a strengthened and timely viral metagenomic detection and routine genomics surveillance. INSaFLU-TELEVIR is compatible with Illumina, Ion Torrent, and ONT data and is freely available at https://insaflu.insa.pt/ (online tool) and https://github.com/INSaFLU (code).
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
INSaFLU: an automated open web-based bioinformatics suite "from-reads" for influenza whole-genome-sequencing-based surveillance.Genome Med. 2018 Jun 29;10(1):46. doi: 10.1186/s13073-018-0555-0. Genome Med. 2018. PMID: 29954441 Free PMC article.
-
A Comparison of Bioinformatics Pipelines for Enrichment Illumina Next Generation Sequencing Systems in Detecting SARS-CoV-2 Virus Strains.Genes (Basel). 2022 Jul 26;13(8):1330. doi: 10.3390/genes13081330. Genes (Basel). 2022. PMID: 35893066 Free PMC article.
-
Entourage: all-in-one sequence analysis software for genome assembly, virus detection, virus discovery, and intrasample variation profiling.BMC Bioinformatics. 2024 Jun 24;25(1):222. doi: 10.1186/s12859-024-05846-y. BMC Bioinformatics. 2024. PMID: 38914932 Free PMC article.
-
Web Resources for Metagenomics Studies.Genomics Proteomics Bioinformatics. 2015 Oct;13(5):296-303. doi: 10.1016/j.gpb.2015.10.003. Epub 2015 Nov 18. Genomics Proteomics Bioinformatics. 2015. PMID: 26602607 Free PMC article. Review.
-
Bioinformatics tools for analysing viral genomic data.Rev Sci Tech. 2016 Apr;35(1):271-85. doi: 10.20506/rst.35.1.2432. Rev Sci Tech. 2016. PMID: 27217183 Review.
Cited by
-
Improving the reporting of metagenomic virome-scale data.Commun Biol. 2024 Dec 20;7(1):1687. doi: 10.1038/s42003-024-07212-3. Commun Biol. 2024. PMID: 39706917 Free PMC article.
-
Molecular and phylogenetic analysis of influenza A and B viruses circulating in Sri Lanka following the COVID-19 pandemic.Virus Genes. 2025 Aug;61(4):444-454. doi: 10.1007/s11262-025-02161-3. Epub 2025 Apr 25. Virus Genes. 2025. PMID: 40279032
-
Preparedness, prevention and control related to zoonotic avian influenza.EFSA J. 2025 Jan 29;23(1):e9191. doi: 10.2903/j.efsa.2025.9191. eCollection 2025 Jan. EFSA J. 2025. PMID: 39882189 Free PMC article.
-
Role of artificial intelligence in advancing immunology.Immunol Res. 2025 Apr 24;73(1):76. doi: 10.1007/s12026-025-09632-7. Immunol Res. 2025. PMID: 40272607 Review.
-
Viral genetics and transmission dynamics in the second wave of mpox outbreak in Portugal and forecasting public health scenarios.Emerg Microbes Infect. 2024 Dec;13(1):2412635. doi: 10.1080/22221751.2024.2412635. Epub 2024 Oct 11. Emerg Microbes Infect. 2024. PMID: 39360827 Free PMC article.
References
-
- European Centre for Disease Prevention and Control (ECDC) Expert opinion on whole genome sequencing for public health surveillance. Stockholm: ECDC; 2016.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

