Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes
- PMID: 38659623
- PMCID: PMC11042495
- DOI: 10.1093/femsml/uqae006
Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes
Abstract
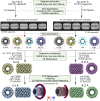
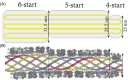
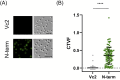
The mimivirus 1.2 Mb genome was shown to be organized into a nucleocapsid-like genomic fiber encased in the nucleoid compartment inside the icosahedral capsid. The genomic fiber protein shell is composed of a mixture of two GMC-oxidoreductase paralogs, one of them being the main component of the glycosylated layer of fibrils at the surface of the virion. In this study, we determined the effect of the deletion of each of the corresponding genes on the genomic fiber and the layer of surface fibrils. First, we deleted the GMC-oxidoreductase, the most abundant in the genomic fiber, and determined its structure and composition in the mutant. As expected, it was composed of the second GMC-oxidoreductase and contained 5- and 6-start helices similar to the wild-type fiber. This result led us to propose a model explaining their coexistence. Then we deleted the GMC-oxidoreductase, the most abundant in the layer of fibrils, to analyze its protein composition in the mutant. Second, we showed that the fitness of single mutants and the double mutant were not decreased compared with the wild-type viruses under laboratory conditions. Third, we determined that deleting the GMC-oxidoreductase genes did not impact the glycosylation or the glycan composition of the layer of surface fibrils, despite modifying their protein composition. Because the glycosylation machinery and glycan composition of members of different clades are different, we expanded the analysis of the protein composition of the layer of fibrils to members of the B and C clades and showed that it was different among the three clades and even among isolates within the same clade. Taken together, the results obtained on two distinct central processes (genome packaging and virion coating) illustrate an unexpected functional redundancy in members of the family Mimiviridae, suggesting this may be the major evolutionary force behind their giant genomes.
Keywords: MS-based proteomics; Mimivirus; cryo-EM; genomic fiber; giant virus; glycosylation; helical reconstruction; layer of fibrils.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.Elife. 2022 Jul 28;11:e77607. doi: 10.7554/eLife.77607. Elife. 2022. PMID: 35900198 Free PMC article.
-
Diversity of Surface Fibril Patterns in Mimivirus Isolates.J Virol. 2023 Feb 28;97(2):e0182422. doi: 10.1128/jvi.01824-22. Epub 2023 Feb 2. J Virol. 2023. PMID: 36728417 Free PMC article.
-
Analyses of the Kroon Virus Major Capsid Gene and Its Transcript Highlight a Distinct Pattern of Gene Evolution and Splicing among Mimiviruses.J Virol. 2018 Jan 2;92(2):e01782-17. doi: 10.1128/JVI.01782-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29118120 Free PMC article.
-
Surface fibrils on the particles of nucleocytoviruses: A review.Exp Biol Med (Maywood). 2023 Nov;248(22):2045-2052. doi: 10.1177/15353702231208410. Epub 2023 Nov 13. Exp Biol Med (Maywood). 2023. PMID: 37955170 Free PMC article. Review.
-
The three-dimensional structure of Mimivirus.Intervirology. 2010;53(5):268-73. doi: 10.1159/000312911. Epub 2010 Jun 15. Intervirology. 2010. PMID: 20551678 Free PMC article. Review.
Cited by
-
Cellular souvenirs in viral luggage.Nat Rev Microbiol. 2025 Jul;23(7):409. doi: 10.1038/s41579-025-01179-6. Nat Rev Microbiol. 2025. PMID: 40204875 No abstract available.
-
20 years of research on giant viruses.Npj Viruses. 2025 Feb 11;3(1):9. doi: 10.1038/s44298-025-00093-1. Npj Viruses. 2025. PMID: 40295850 Free PMC article. Review.
-
Melbournevirus encodes a shorter H2B-H2A doublet histone variant that forms structurally distinct nucleosome structures.Nat Commun. 2025 Jul 26;16(1):6903. doi: 10.1038/s41467-025-62031-2. Nat Commun. 2025. PMID: 40715086 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
