Crystal Polymorph Search in the NPT Ensemble via a Deposition/Sublimation Alchemical Path
- PMID: 38659664
- PMCID: PMC11036363
- DOI: 10.1021/acs.cgd.3c01358
Crystal Polymorph Search in the NPT Ensemble via a Deposition/Sublimation Alchemical Path
Abstract
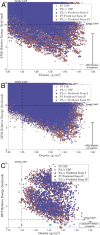
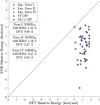
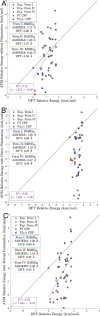
The formulation of active pharmaceutical ingredients involves discovering stable crystal packing arrangements or polymorphs, each of which has distinct pharmaceutically relevant properties. Traditional experimental screening techniques utilizing various conditions are commonly supplemented with in silico crystal structure prediction (CSP) to inform the crystallization process and mitigate risk. Predictions are often based on advanced classical force fields or quantum mechanical calculations that model the crystal potential energy landscape but do not fully incorporate temperature, pressure, or solution conditions during the search procedure. This study proposes an innovative alchemical path that utilizes an advanced polarizable atomic multipole force field to predict crystal structures based on direct sampling of the NPT ensemble. The use of alchemical (i.e., nonphysical) intermediates, a novel Monte Carlo barostat, and an orthogonal space tempering bias combine to enhance the sampling efficiency of the deposition/sublimation phase transition. The proposed algorithm was applied to 2-((4-(2-(3,4-dichlorophenyl)ethyl)phenyl)amino)benzoic acid (Cambridge Crystallography Database Centre ID: XAFPAY) as a case study to showcase the algorithm. Each experimentally determined polymorph with one molecule in the asymmetric unit was successfully reproduced via approximately 1000 short 1 ns simulations per space group where each simulation was initiated from random rigid body coordinates and unit cell parameters. Utilizing two threads of a recent Intel CPU (a Xeon Gold 6330 CPU at 2.00 GHz), 1 ns of sampling using the polarizable AMOEBA force field can be acquired in 4 h (equating to more than 300 ns/day using all 112 threads/56 cores of a dual CPU node) within the Force Field X software (https://ffx.biochem.uiowa.edu). These results demonstrate a step forward in the rigorous use of the NPT ensemble during the CSP search process and open the door to future algorithms that incorporate solution conditions using continuum solvation methods.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





Similar articles
-
Toward polarizable AMOEBA thermodynamics at fixed charge efficiency using a dual force field approach: application to organic crystals.Phys Chem Chem Phys. 2016 Nov 9;18(44):30313-30322. doi: 10.1039/c6cp02595a. Phys Chem Chem Phys. 2016. PMID: 27524378 Free PMC article.
-
Polymorph sampling with coupling to extended variables: enhanced sampling of polymorph energy landscapes and free energy perturbation of polymorph ensembles.Acta Crystallogr B Struct Sci Cryst Eng Mater. 2024 Dec 1;80(Pt 6):575-94. doi: 10.1107/S205252062400132X. Online ahead of print. Acta Crystallogr B Struct Sci Cryst Eng Mater. 2024. PMID: 39405193 Free PMC article.
-
Polarizable Atomic Multipole X-Ray Refinement: Particle Mesh Ewald Electrostatics for Macromolecular Crystals.J Chem Theory Comput. 2011 Apr 12;7(4):1141-56. doi: 10.1021/ct100506d. Epub 2011 Mar 9. J Chem Theory Comput. 2011. PMID: 26606362
-
Unprecedented Packing Polymorphism of Oxindole: An Exploration Inspired by Crystal Structure Prediction.Angew Chem Int Ed Engl. 2024 Aug 19;63(34):e202406214. doi: 10.1002/anie.202406214. Epub 2024 Jul 17. Angew Chem Int Ed Engl. 2024. PMID: 38825853
-
From crystal structure prediction to polymorph prediction: interpreting the crystal energy landscape.Phys Chem Chem Phys. 2008 Apr 21;10(15):1996-2009. doi: 10.1039/b719351c. Epub 2008 Feb 19. Phys Chem Chem Phys. 2008. PMID: 18688351 Review.
Cited by
-
Force Field X: A computational microscope to study genetic variation and organic crystals using theory and experiment.J Chem Phys. 2024 Jul 7;161(1):012501. doi: 10.1063/5.0214652. J Chem Phys. 2024. PMID: 38958156 Free PMC article.
References
-
- Brittain H. G.Polymorphism in Pharmaceutical Solids; CRC Press, 2018.
-
- Bernstein J.Polymorphism in Molecular Crystals; Oxford University Press, 2020.
-
- Hilfiker R.; von Raumer M.. Polymorphism in the Pharmaceutical Industry: Solid Form and Drug Development; Wiley, 2019.
LinkOut - more resources
Full Text Sources
Miscellaneous
