This is a preprint.
Adam19 Deficiency Impacts Pulmonary Function: Human GWAS Follow-up in Mouse
- PMID: 38659817
- PMCID: PMC11042436
- DOI: 10.21203/rs.3.rs-4207678/v1
Adam19 Deficiency Impacts Pulmonary Function: Human GWAS Follow-up in Mouse
Update in
-
Adam19 Deficiency Impacts Pulmonary Function: Human GWAS Follow-up in a Mouse Knockout Model.Lung. 2024 Oct;202(5):659-672. doi: 10.1007/s00408-024-00738-7. Epub 2024 Aug 17. Lung. 2024. PMID: 39153120 Free PMC article.
Abstract
Purpose: Over 550 loci have been associated with human pulmonary function in genome-wide association studies (GWAS); however, the causal role of most remains uncertain. Single nucleotide polymorphisms in a disintegrin and metalloprotease domain 19 (ADAM19) are consistently related to pulmonary function in GWAS. Thus, we used a mouse model to investigate the causal link between Adam19 and pulmonary function.
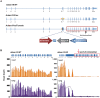
Methods: We created an Adam19 knockout (KO) mouse model and validated the gene targeting using RNA-Seq and RT-qPCR. Contrary to prior publications, the KO was not neonatal lethal. Thus, we phenotyped the Adam19 KO.
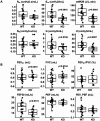
Results: KO mice had lower body weight and shorter tibial length than wild type (WT). Dual-energy X-ray Absorptiometry indicated lower soft weight, fat weight, and bone mineral content in KO mice. In lung function analyses using flexiVent, compared to WT, Adam19 KO had decreased baseline respiratory system elastance, minute work of breathing, tissue damping, tissue elastance, and forced expiratory flow at 50% forced vital capacity but higher FEV0.1 and FVC. Adam19 KO had attenuated tissue damping and tissue elastance in response to methacholine following LPS exposure. Adam19 KO also exhibited attenuated neutrophil extravasation into the airway after LPS administration compared to WT. RNA-Seq analysis of KO and WT lungs identified several differentially expressed genes (Cd300lg, Kpna2, and Pttg1) implicated in lung biology and pathogenesis. Gene set enrichment analysis identified negative enrichment for TNF pathways.
Conclusion: Our murine findings support a causal role of ADAM19, implicated in human GWAS, in regulating pulmonary function.
Keywords: RNA-Seq; flexiVent; inflammation; lung function; meltrin beta; spirometry.
Conflict of interest statement
Competing Interests The authors have no relevant financial or non-financial interests to disclose.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

