This is a preprint.
PAbFold: Linear Antibody Epitope Prediction using AlphaFold2
- PMID: 38659833
- PMCID: PMC11042291
- DOI: 10.1101/2024.04.19.590298
PAbFold: Linear Antibody Epitope Prediction using AlphaFold2
Abstract
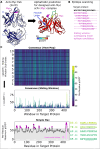
Defining the binding epitopes of antibodies is essential for understanding how they bind to their antigens and perform their molecular functions. However, while determining linear epitopes of monoclonal antibodies can be accomplished utilizing well-established empirical procedures, these approaches are generally labor- and time-intensive and costly. To take advantage of the recent advances in protein structure prediction algorithms available to the scientific community, we developed a calculation pipeline based on the localColabFold implementation of AlphaFold2 that can predict linear antibody epitopes by predicting the structure of the complex between antibody heavy and light chains and target peptide sequences derived from antigens. We found that this AlphaFold2 pipeline, which we call PAbFold, was able to accurately flag known epitope sequences for several well-known antibody targets (HA / Myc) when the target sequence was broken into small overlapping linear peptides and antibody complementarity determining regions (CDRs) were grafted onto several different antibody framework regions in the single-chain antibody fragment (scFv) format. To determine if this pipeline was able to identify the epitope of a novel antibody with no structural information publicly available, we determined the epitope of a novel anti-SARS-CoV-2 nucleocapsid targeted antibody using our method and then experimentally validated our computational results using peptide competition ELISA assays. These results indicate that the AlphaFold2-based PAbFold pipeline we developed is capable of accurately identifying linear antibody epitopes in a short time using just antibody and target protein sequences. This emergent capability of the method is sensitive to methodological details such as peptide length, AlphaFold2 neural network versions, and multiple-sequence alignment database. PAbFold is available at https://github.com/jbderoo/PAbFold.
Keywords: AlphaFold2; antibody; competition ELISA; epitope-prediction; linear epitope; scFv.
Figures



References
-
- Saha S, Raghava GPS. 2006. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 65:40–8. - PubMed
-
- Ambrosetti F, Jiménez-García B, Roel-Touris J, Bonvin AMJJ. 2020. Modeling Antibody-Antigen Complexes by Information-Driven Docking. Structure 28:119–129.e2. - PubMed
-
- Dominguez C, Boelens R, Bonvin AMJJ. 2003. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125:1731–1737. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
