The acetylation of MDH1 and IDH1 is associated with energy metabolism in acute liver failure
- PMID: 38660411
- PMCID: PMC11039345
- DOI: 10.1016/j.isci.2024.109678
The acetylation of MDH1 and IDH1 is associated with energy metabolism in acute liver failure
Abstract
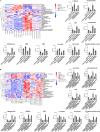
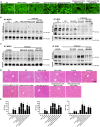
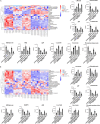
The liver is the main organ associated with metabolism. In our previous studies, we identified that the metabolic enzymes malate dehydrogenase 1 (MDH1) and isocitrate dehydrogenase 1 (IDH1) were differentially expressed in ALF. The aim of this study was to explore the changes in the acetylation of MDH1 and IDH1 and the therapeutic effect of histone deacetylase (HDAC) inhibitor in acute liver failure (ALF). Decreased levels of many metabolites were observed in ALF patients. MDH1 and IDH1 were decreased in the livers of ALF patients. The HDAC inhibitor ACY1215 improved the expression of MDH1 and IDH1 after treatment with MDH1-siRNA and IDH1-siRNA. Transfection with mutant plasmids and adeno-associated viruses, identified MDH1 K118 acetylation and IDH1 K93 acetylation as two important sites that regulate metabolism in vitro and in vivo.
Keywords: Hepatology; Human metabolism; Molecular biology.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no competing of interests.
Figures

Similar articles
-
Deacetylated MDH1 and IDH1 aggravates PANoptosis in acute liver failure through endoplasmic reticulum stress signaling.Cell Death Discov. 2024 Jun 8;10(1):275. doi: 10.1038/s41420-024-02054-8. Cell Death Discov. 2024. PMID: 38851781 Free PMC article.
-
IDH1/MDH1 deacetylation promotes NETosis by regulating OPA1 and autophagy.Int Immunopharmacol. 2024 Dec 25;143(Pt 1):113270. doi: 10.1016/j.intimp.2024.113270. Epub 2024 Sep 30. Int Immunopharmacol. 2024. PMID: 39353390
-
IDH1/MDH1 deacetylation promotes acute liver failure by regulating NETosis.Cell Mol Biol Lett. 2024 Jan 3;29(1):8. doi: 10.1186/s11658-023-00529-7. Cell Mol Biol Lett. 2024. PMID: 38172700 Free PMC article.
-
Role of malate dehydrogenase 1 and isocitrate dehydrogenase 1 and their posttranslational modifications in diseases.Biochem Biophys Res Commun. 2025 Mar 25;754:151535. doi: 10.1016/j.bbrc.2025.151535. Epub 2025 Feb 24. Biochem Biophys Res Commun. 2025. PMID: 40022816 Review.
-
[Epigenetic mechanisms and alcohol use disorders: a potential therapeutic target].Biol Aujourdhui. 2017;211(1):83-91. doi: 10.1051/jbio/2017014. Epub 2017 Jul 6. Biol Aujourdhui. 2017. PMID: 28682229 Review. French.
Cited by
-
Putative biomarkers of hepatic dysfunction in critically ill sepsis patients.Clin Exp Med. 2025 Jan 3;25(1):28. doi: 10.1007/s10238-024-01545-3. Clin Exp Med. 2025. PMID: 39751971 Free PMC article.
-
Deacetylated MDH1 and IDH1 aggravates PANoptosis in acute liver failure through endoplasmic reticulum stress signaling.Cell Death Discov. 2024 Jun 8;10(1):275. doi: 10.1038/s41420-024-02054-8. Cell Death Discov. 2024. PMID: 38851781 Free PMC article.
-
Mechanisms of sepsis-induced acute liver injury: a comprehensive review.Front Cell Infect Microbiol. 2025 Feb 21;15:1504223. doi: 10.3389/fcimb.2025.1504223. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40061452 Free PMC article. Review.
References
-
- Nishikawa T., Bellance N., Damm A., Bing H., Zhu Z., Handa K., Yovchev M.I., Sehgal V., Moss T.J., Oertel M., et al. A switch in the source of ATP production and a loss in capacity to perform glycolysis are hallmarks of hepatocyte failure in advance liver disease. J. Hepatol. 2014;60:1203–1211. doi: 10.1016/j.jhep.2014.02.014. - DOI - PMC - PubMed
-
- Meng Q.H., Hou W., Yu H.W., Lu J., Li J., Wang J.H., Zhang F.Y., Zhang J., Yao Q.W., Wu J., et al. Resting energy expenditure and substrate metabolism in patients with acute-on-chronic hepatitis B liver failure. J. Clin. Gastroenterol. 2011;45:456–461. doi: 10.1097/MCG.0b013e31820f7f02. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

