Exploring the roles and potential therapeutic strategies of inflammation and metabolism in the pathogenesis of vitiligo: a mendelian randomization and bioinformatics-based investigation
- PMID: 38660673
- PMCID: PMC11039897
- DOI: 10.3389/fgene.2024.1385339
Exploring the roles and potential therapeutic strategies of inflammation and metabolism in the pathogenesis of vitiligo: a mendelian randomization and bioinformatics-based investigation
Abstract
Introduction: Vitiligo, a common autoimmune acquired pigmentary skin disorder, poses challenges due to its unclear pathogenesis. Evidence suggests inflammation and metabolism's pivotal roles in its onset and progression. This study aims to elucidate the causal relationships between vitiligo and inflammatory proteins, immune cells, and metabolites, exploring bidirectional associations and potential drug targets.
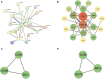
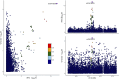
Methods: Mendelian Randomization (MR) analysis encompassed 4,907 plasma proteins, 91 inflammatory proteins, 731 immune cell features, and 1400 metabolites. Bioinformatics analysis included Protein-Protein Interaction (PPI) network construction, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Subnetwork discovery and hub protein identification utilized the Molecular Complex Detection (MCODE) plugin. Colocalization analysis and drug target exploration, including molecular docking validation, were performed.
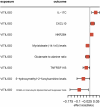
Results: MR analysis identified 49 proteins, 39 immune cell features, and 59 metabolites causally related to vitiligo. Bioinformatics analysis revealed significant involvement in PPI, GO enrichment, and KEGG pathways. Subnetwork analysis identified six central proteins, with Interferon Regulatory Factor 3 (IRF3) exhibiting strong colocalization evidence. Molecular docking validated Piceatannol's binding to IRF3, indicating a stable interaction.
Conclusion: This study comprehensively elucidates inflammation, immune response, and metabolism's intricate involvement in vitiligo pathogenesis. Identified proteins and pathways offer potential therapeutic targets, with IRF3 emerging as a promising candidate. These findings deepen our understanding of vitiligo's etiology, informing future research and drug development endeavors.
Keywords: bioinformatics; inflammation; mendelian randomization; metabolism; therapeutic strategies; vitiligo.
Copyright © 2024 He, Ran, Zhang, Fu, He, Zhang, Mao, Zhao, Yin and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

