Probing the salivary proteome for prognostic biomarkers in response to non-surgical periodontal therapy
- PMID: 38660744
- PMCID: PMC11671166
- DOI: 10.1111/jcpe.13990
Probing the salivary proteome for prognostic biomarkers in response to non-surgical periodontal therapy
Abstract
Aim: This prospective study investigated the salivary proteome before and after periodontal therapy.
Materials and methods: Ten systemically healthy, non-smoking, stage III, grade C periodontitis patients underwent non-surgical periodontal treatment. Full-mouth periodontal parameters were measured, and saliva (n = 30) collected pre- (T0), and one (T1) and six (T6) months post-treatment. The proteome was investigated by label-free quantitative proteomics. Protein expression changes were modelled over time, with significant protein regulation considered at false discovery rate <0.05.
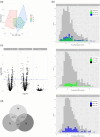
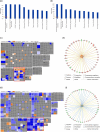
Results: Treatment significantly reduced bleeding scores, percentages of sites with pocket depth ≥5 mm, plaque and gingival indexes. One thousand seven hundred and thirteen proteins were identified and 838 proteins (human = 757, bacterial = 81) quantified (≥2 peptides). At T1, 80 (T1 vs. T0: 60↑:20↓), and at T6, 118 human proteins (T6 vs. T0: 67↑:51↓) were regulated. The salivary proteome at T6 versus T1 remained stable. Highest protein activity post- versus pre-treatment was observed for cellular movement and inflammatory response. The small proline-rich protein 3 (T1 vs. T0: 5.4-fold↑) and lymphocyte-specific protein 1 (T6 vs. T0: 4.6-fold↓) were the top regulated human proteins. Proteins from Neisseria mucosa and Treponema socranskii (T1 vs. T0: 8.0-fold↓, 4.9-fold↓) were down-regulated.
Conclusions: Periodontal treatment reduced clinical disease parameters and these changes were reflected in the salivary proteome. This underscores the potential of utilizing saliva biomarkers as prognostic tools for monitoring treatment outcomes.
Keywords: biomarkers; label‐free quantitative proteomics; periodontal treatment; periodontitis; saliva; subgingival instrumentation.
© 2024 The Authors. Journal of Clinical Periodontology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Baumgartner, D. , Johannsen, B. , Specht, M. , Lüddecke, J. , Rombach, M. , Hin, S. , Paust, N. , von Stetten, F. , Zengerle, R. , Herz, C. , Peham, J. R. , Paqué, P. N. , Attin, T. , Jenzer, J. S. , Körner, P. , Schmidlin, P. R. , Thurnheer, T. , Wegehaupt, F. J. , Kaman, W. E. , … Mitsakakis, K. (2021). OralDisk: A chair‐side compatible molecular platform using whole saliva for monitoring oral health at the dental practice. Biosensors, 11(11), 423. 10.3390/bios11110423 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

