SPTLC3 Is Essential for Complex I Activity and Contributes to Ischemic Cardiomyopathy
- PMID: 38660786
- PMCID: PMC11333184
- DOI: 10.1161/CIRCULATIONAHA.123.066879
SPTLC3 Is Essential for Complex I Activity and Contributes to Ischemic Cardiomyopathy
Abstract
Background: Dysregulated metabolism of bioactive sphingolipids, including ceramides and sphingosine-1-phosphate, has been implicated in cardiovascular disease, although the specific species, disease contexts, and cellular roles are not completely understood. Sphingolipids are produced by the serine palmitoyltransferase enzyme, canonically composed of 2 subunits, SPTLC1 (serine palmitoyltransferase long chain base subunit 1) and SPTLC2 (serine palmitoyltransferase long chain base subunit 2). Noncanonical sphingolipids are produced by a more recently described subunit, SPTLC3 (serine palmitoyltransferase long chain base subunit 3).
Methods: The noncanonical (d16) and canonical (d18) sphingolipidome profiles in cardiac tissues of patients with end-stage ischemic cardiomyopathy and in mice with ischemic cardiomyopathy were analyzed by targeted lipidomics. Regulation of SPTLC3 by HIF1α under ischemic conditions was determined with chromatin immunoprecipitation. Transcriptomics, lipidomics, metabolomics, echocardiography, mitochondrial electron transport chain, mitochondrial membrane fluidity, and mitochondrial membrane potential were assessed in the cSPTLC3KO transgenic mice we generated. Furthermore, morphological and functional studies were performed on cSPTLC3KO mice subjected to permanent nonreperfused myocardial infarction.
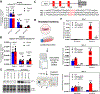
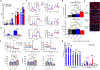
Results: Herein, we report that SPTLC3 is induced in both human and mouse models of ischemic cardiomyopathy and leads to production of atypical sphingolipids bearing 16-carbon sphingoid bases, resulting in broad changes in cell sphingolipid composition. This induction is in part attributable to transcriptional regulation by HIF1α under ischemic conditions. Furthermore, cardiomyocyte-specific depletion of SPTLC3 in mice attenuates oxidative stress, fibrosis, and hypertrophy in chronic ischemia, and mice demonstrate improved cardiac function and increased survival along with increased ketone and glucose substrate metabolism utilization. Depletion of SPTLC3 mechanistically alters the membrane environment and subunit composition of mitochondrial complex I of the electron transport chain, decreasing its activity.
Conclusions: Our findings suggest a novel essential role for SPTLC3 in electron transport chain function and a contribution to ischemic injury by regulating complex I activity.
Keywords: cardiomyopathy; electron transport complex I; mitochondria; serine C-palmitoyltransferase; sphingolipids.
Conflict of interest statement
None.
Figures




References
-
- Heidenreich PA, Bozkurt B, Aguilar D, Allen LA, Byun JJ, Colvin MM, et al. 2022 AHA/ACC/HFSA guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Journal of the American College of Cardiology. 2022;79(17):e263–e421. - PubMed
-
- Nowbar AN, Gitto M, Howard JP, Francis DP, and Al-Lamee R. Mortality from ischemic heart disease: Analysis of data from the World Health Organization and coronary artery disease risk factors From NCD Risk Factor Collaboration. Circulation: cardiovascular quality and outcomes. 2019;12(6):e005375. - PMC - PubMed
-
- Beatty AL, Ku IA, Bibbins‐Domingo K, Christenson RH, DeFilippi CR, Ganz P, et al. Traditional risk factors versus biomarkers for prediction of secondary events in patients with stable coronary heart disease: from the heart and soul study. Journal of the American Heart Association. 2015;4(7):e001646. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- R01 HL151243/HL/NHLBI NIH HHS/United States
- I01 BX003725/BX/BLRD VA/United States
- I21 RX004398/RX/RRD VA/United States
- R56 DK097907/DK/NIDDK NIH HHS/United States
- K08 HL155852/HL/NHLBI NIH HHS/United States
- R01 HL117233/HL/NHLBI NIH HHS/United States
- R21 AA029518/AA/NIAAA NIH HHS/United States
- I01 BX001963/BX/BLRD VA/United States
- F31 HL156529/HL/NHLBI NIH HHS/United States
- S10 OD023527/OD/NIH HHS/United States
- IK6 BX006315/BX/BLRD VA/United States
- I01 BX001355/BX/BLRD VA/United States
- I01 BX003429/BX/BLRD VA/United States
- I01 BX000200/BX/BLRD VA/United States
- R01 HL144776/HL/NHLBI NIH HHS/United States
- R35 HL155651/HL/NHLBI NIH HHS/United States
- P30 CA016059/CA/NCI NIH HHS/United States
- T32 HL149645/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

