Mild Hypoxia Accelerates Cerebral Cavernous Malformation Disease Through CX3CR1-CX3CL1 Signaling
- PMID: 38660801
- PMCID: PMC11111348
- DOI: 10.1161/ATVBAHA.123.320367
Mild Hypoxia Accelerates Cerebral Cavernous Malformation Disease Through CX3CR1-CX3CL1 Signaling
Abstract
Background: Heterogeneity in the severity of cerebral cavernous malformations (CCMs) disease, including brain bleedings and thrombosis that cause neurological disabilities in patients, suggests that environmental, genetic, or biological factors act as disease modifiers. Still, the underlying mechanisms are not entirely understood. Here, we report that mild hypoxia accelerates CCM disease by promoting angiogenesis, neuroinflammation, and vascular thrombosis in the brains of CCM mouse models.
Methods: We used genetic studies, RNA sequencing, spatial transcriptome, micro-computed tomography, fluorescence-activated cell sorting, multiplex immunofluorescence, coculture studies, and imaging techniques to reveal that sustained mild hypoxia via the CX3CR1-CX3CL1 (CX3C motif chemokine receptor 1/chemokine [CX3C motif] ligand 1) signaling pathway influences cell-specific neuroinflammatory interactions, contributing to heterogeneity in CCM severity.
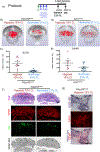
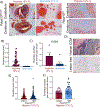
Results: Histological and expression profiles of CCM neurovascular lesions (Slco1c1-iCreERT2;Pdcd10fl/fl; Pdcd10BECKO) in male and female mice found that sustained mild hypoxia (12% O2, 7 days) accelerates CCM disease. Our findings indicate that a small reduction in oxygen levels can significantly increase angiogenesis, neuroinflammation, and thrombosis in CCM disease by enhancing the interactions between endothelium, astrocytes, and immune cells. Our study indicates that the interactions between CX3CR1 and CX3CL1 are crucial in the maturation of CCM lesions and propensity to CCM immunothrombosis. In particular, this pathway regulates the recruitment and activation of microglia and other immune cells in CCM lesions, which leads to lesion growth and thrombosis. We found that human CX3CR1 variants are linked to lower lesion burden in familial CCMs, proving it is a genetic modifier in human disease and a potential marker for aggressiveness. Moreover, monoclonal blocking antibody against CX3CL1 or reducing 1 copy of the Cx3cr1 gene significantly reduces hypoxia-induced CCM immunothrombosis.
Conclusions: Our study reveals that interactions between CX3CR1 and CX3CL1 can modify CCM neuropathology when lesions are accelerated by environmental hypoxia. Moreover, a hypoxic environment or hypoxia signaling caused by CCM disease influences the balance between neuroinflammation and neuroprotection mediated by CX3CR1-CX3CL1 signaling. These results establish CX3CR1 as a genetic marker for patient stratification and a potential predictor of CCM aggressiveness.
Keywords: genetic markers; hemangioma, cavernous, central nervous system; hypoxia; neuroinflammatory diseases; thromboinflammation.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

