Dual fluorescence reporter mice for Ccl3 transcription, translation, and intercellular communication
- PMID: 38661718
- PMCID: PMC11044946
- DOI: 10.1084/jem.20231814
Dual fluorescence reporter mice for Ccl3 transcription, translation, and intercellular communication
Abstract
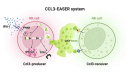
Chemokines guide immune cells during their response against pathogens and tumors. Various techniques exist to determine chemokine production, but none to identify cells that directly sense chemokines in vivo. We have generated CCL3-EASER (ErAse, SEnd, Receive) mice that simultaneously report for Ccl3 transcription and translation, allow identifying Ccl3-sensing cells, and permit inducible deletion of Ccl3-producing cells. We infected these mice with murine cytomegalovirus (mCMV), where Ccl3 and NK cells are critical defense mediators. We found that NK cells transcribed Ccl3 already in homeostasis, but Ccl3 translation required type I interferon signaling in infected organs during early infection. NK cells were both the principal Ccl3 producers and sensors of Ccl3, indicating auto/paracrine communication that amplified NK cell response, and this was essential for the early defense against mCMV. CCL3-EASER mice represent the prototype of a new class of dual fluorescence reporter mice for analyzing cellular communication via chemokines, which may be applied also to other chemokines and disease models.
© 2024 Rodrigo et al.
Conflict of interest statement
Disclosures: C. Martin-Higueras reported personal fees from Novo Nordisk and Arbor Biotechnologies outside the submitted work. No other disclosures were reported.
Figures











References
-
- Bekiaris, V., and Lane P.J.L.. 2010. The localization and migration of natural killer cells in health and disease. Natural Killer Cells. 137–153., 10.1016/B978-0-12-370454-2.00010-7 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

