Cryo-EM structures of membrane-bound dynamin in a post-hydrolysis state primed for membrane fission
- PMID: 38663399
- PMCID: PMC11265984
- DOI: 10.1016/j.devcel.2024.04.008
Cryo-EM structures of membrane-bound dynamin in a post-hydrolysis state primed for membrane fission
Abstract
Dynamin assembles as a helical polymer at the neck of budding endocytic vesicles, constricting the underlying membrane as it progresses through the GTPase cycle to sever vesicles from the plasma membrane. Although atomic models of the dynamin helical polymer bound to guanosine triphosphate (GTP) analogs define earlier stages of membrane constriction, there are no atomic models of the assembled state post-GTP hydrolysis. Here, we used cryo-EM methods to determine atomic structures of the dynamin helical polymer assembled on lipid tubules, akin to necks of budding endocytic vesicles, in a guanosine diphosphate (GDP)-bound, super-constricted state. In this state, dynamin is assembled as a 2-start helix with an inner lumen of 3.4 nm, primed for spontaneous fission. Additionally, by cryo-electron tomography, we trapped dynamin helical assemblies within HeLa cells using the GTPase-defective dynamin K44A mutant and observed diverse dynamin helices, demonstrating that dynamin can accommodate a range of assembled complexes in cells that likely precede membrane fission.
Keywords: clathrin-mediated endocytosis; cryo-EM; cryo-ET; dynamin; endocytosis; membrane fission; membrane remodeling; structural biology.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



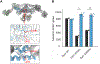
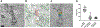

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

