Numb positively regulates Hedgehog signaling at the ciliary pocket
- PMID: 38664376
- PMCID: PMC11045789
- DOI: 10.1038/s41467-024-47244-1
Numb positively regulates Hedgehog signaling at the ciliary pocket
Abstract
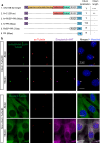
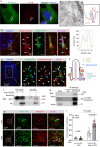
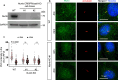
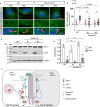
Hedgehog (Hh) signaling relies on the primary cilium, a cell surface organelle that serves as a signaling hub for the cell. Using proximity labeling and quantitative proteomics, we identify Numb as a ciliary protein that positively regulates Hh signaling. Numb localizes to the ciliary pocket and acts as an endocytic adaptor to incorporate Ptch1 into clathrin-coated vesicles, thereby promoting Ptch1 exit from the cilium, a key step in Hh signaling activation. Numb loss impedes Sonic hedgehog (Shh)-induced Ptch1 exit from the cilium, resulting in reduced Hh signaling. Numb loss in spinal neural progenitors reduces Shh-induced differentiation into cell fates reliant on high Hh activity. Genetic ablation of Numb in the developing cerebellum impairs the proliferation of granule cell precursors, a Hh-dependent process, resulting in reduced cerebellar size. This study highlights Numb as a regulator of ciliary Ptch1 levels during Hh signal activation and demonstrates the key role of ciliary pocket-mediated endocytosis in cell signaling.
© 2024. The Author(s).
Conflict of interest statement
Tess C. Branon is an employee of Interline Therapeutics, South San Francisco, CA, USA. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

