Photobody formation spatially segregates two opposing phytochrome B signaling actions of PIF5 degradation and stabilization
- PMID: 38664420
- PMCID: PMC11045832
- DOI: 10.1038/s41467-024-47790-8
Photobody formation spatially segregates two opposing phytochrome B signaling actions of PIF5 degradation and stabilization
Abstract
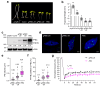
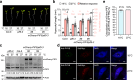
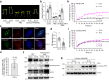
Photoactivation of the plant photoreceptor and thermosensor phytochrome B (PHYB) triggers its condensation into subnuclear membraneless organelles named photobodies (PBs). However, the function of PBs in PHYB signaling remains frustratingly elusive. Here, we found that PHYB recruits PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) to PBs. Surprisingly, PHYB exerts opposing roles in degrading and stabilizing PIF5. Perturbing PB size by overproducing PHYB provoked a biphasic PIF5 response: while a moderate increase in PHYB enhanced PIF5 degradation, further elevating the PHYB level stabilized PIF5 by retaining more of it in enlarged PBs. Conversely, reducing PB size by dim light, which enhanced PB dynamics and nucleoplasmic PHYB and PIF5, switched the balance towards PIF5 degradation. Together, these results reveal that PB formation spatially segregates two antagonistic PHYB signaling actions - PIF5 stabilization in PBs and PIF5 degradation in the surrounding nucleoplasm - which could enable an environmentally sensitive, counterbalancing mechanism to titrate nucleoplasmic PIF5 and environmental responses.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
Photobody formation spatially segregates two opposing phytochrome B signaling actions to titrate plant environmental responses.bioRxiv [Preprint]. 2024 Jan 5:2023.11.12.566724. doi: 10.1101/2023.11.12.566724. bioRxiv. 2024. Update in: Nat Commun. 2024 Apr 25;15(1):3519. doi: 10.1038/s41467-024-47790-8. PMID: 38014306 Free PMC article. Updated. Preprint.

