Gut microbiome composition reveals the distinctiveness between the Bengali people and the Indigenous ethnicities in Bangladesh
- PMID: 38664512
- PMCID: PMC11045797
- DOI: 10.1038/s42003-024-06191-9
Gut microbiome composition reveals the distinctiveness between the Bengali people and the Indigenous ethnicities in Bangladesh
Abstract
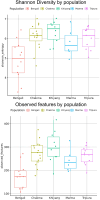
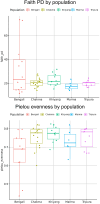
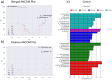
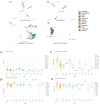
Ethnicity has a significant role in shaping the composition of the gut microbiome, which has implications in human physiology. This study intends to investigate the gut microbiome of Bengali people as well as several indigenous ethnicities (Chakma, Marma, Khyang, and Tripura) residing in the Chittagong Hill Tracts areas of Bangladesh. Following fecal sample collection from each population, part of the bacterial 16 s rRNA gene was amplified and sequenced using Illumina NovaSeq platform. Our findings indicated that Bangladeshi gut microbiota have a distinct diversity profile when compared to other countries. We also found out that Bangladeshi indigenous communities had a higher Firmicutes to Bacteroidetes ratio than the Bengali population. The investigation revealed an unclassified bacterium that was differentially abundant in Bengali samples while the genus Alistipes was found to be prevalent in Chakma samples. Further research on these bacteria might help understand diseases associated with these populations. Also, the current small sample-sized pilot study hindered the comprehensive understanding of the gut microbial diversity of the Bangladeshi population and its potential health implications. However, our study will help establish a basic understanding of the gut microbiome of the Bangladeshi population.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Genetic hypogonadal mouse model reveals niche-specific influence of reproductive axis and sex on intestinal microbial communities.Biol Sex Differ. 2023 Nov 6;14(1):79. doi: 10.1186/s13293-023-00564-1. Biol Sex Differ. 2023. PMID: 37932822 Free PMC article.
-
Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States.PLoS One. 2013;8(1):e53838. doi: 10.1371/journal.pone.0053838. Epub 2013 Jan 22. PLoS One. 2013. PMID: 23349750 Free PMC article.
-
The Association Between Smoking and Gut Microbiome in Bangladesh.Nicotine Tob Res. 2020 Jul 16;22(8):1339-1346. doi: 10.1093/ntr/ntz220. Nicotine Tob Res. 2020. PMID: 31794002 Free PMC article.
-
Impact of urbanization on gut microbiome mosaics across geographic and dietary contexts.mSystems. 2024 Oct 22;9(10):e0058524. doi: 10.1128/msystems.00585-24. Epub 2024 Sep 17. mSystems. 2024. PMID: 39287374 Free PMC article.
-
The association between gut microbiome and anthropometric measurements in Bangladesh.Gut Microbes. 2020;11(1):63-76. doi: 10.1080/19490976.2019.1614394. Epub 2019 May 29. Gut Microbes. 2020. PMID: 31138061 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

