Extensive protein pyrophosphorylation revealed in human cell lines
- PMID: 38664588
- PMCID: PMC11427299
- DOI: 10.1038/s41589-024-01613-5
Extensive protein pyrophosphorylation revealed in human cell lines
Abstract
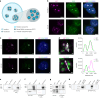
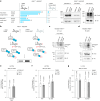
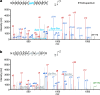
Reversible protein phosphorylation is a central signaling mechanism in eukaryotes. Although mass-spectrometry-based phosphoproteomics has become routine, identification of non-canonical phosphorylation has remained a challenge. Here we report a tailored workflow to detect and reliably assign protein pyrophosphorylation in two human cell lines, providing, to our knowledge, the first direct evidence of endogenous protein pyrophosphorylation. We manually validated 148 pyrophosphosites across 71 human proteins, the most heavily pyrophosphorylated of which were the nucleolar proteins NOLC1 and TCOF1. Detection was consistent with previous biochemical evidence relating the installation of the modification to inositol pyrophosphates (PP-InsPs). When the biosynthesis of PP-InsPs was perturbed, proteins expressed in this background exhibited no signs of pyrophosphorylation. Disruption of PP-InsP biosynthesis also significantly reduced rDNA transcription, potentially by lowering pyrophosphorylation on regulatory proteins NOLC1, TCOF1 and UBF1. Overall, protein pyrophosphorylation emerges as an archetype of non-canonical phosphorylation and should be considered in future phosphoproteomic analyses.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Cohen, P. The origins of protein phosphorylation. Nat. Cell Biol.4, E127–E130 (2002). - PubMed
-
- Humphrey, S. J., James, D. E. & Mann, M. Protein phosphorylation: a major switch mechanism for metabolic regulation. Trends Endocrinol. Metab.26, 676–687 (2015). - PubMed
-
- Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science298, 1912–1934 (2002). - PubMed
-
- Alonso, A. et al. Protein tyrosine phosphatases in the human genome. Cell117, 699–711 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

