Thyroid hormone induces ossification and terminal maturation in a preserved OA cartilage biomimetic model
- PMID: 38664820
- PMCID: PMC11044551
- DOI: 10.1186/s13075-024-03326-5
Thyroid hormone induces ossification and terminal maturation in a preserved OA cartilage biomimetic model
Abstract
Objective: To characterize aspects of triiodothyronine (T3) induced chondrocyte terminal maturation within the molecular osteoarthritis pathophysiology using the previously established T3 human ex vivo osteochondral explant model.
Designs: RNA-sequencing was performed on explant cartilage obtained from OA patients (n = 8), that was cultured ex vivo with or without T3 (10 ng/ml), and main findings were validated using RT-qPCR in an independent sample set (n = 22). Enrichment analysis was used for functional clustering and comparisons with available OA patient RNA-sequencing and GWAS datasets were used to establish relevance for OA pathophysiology by linking to OA patient genomic profiles.
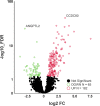
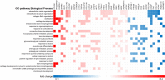
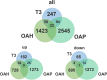
Results: Besides the upregulation of known hypertrophic genes EPAS1 and ANKH, T3 treatment resulted in differential expression of 247 genes with main pathways linked to extracellular matrix and ossification. CCDC80, CDON, ANKH and ATOH8 were among the genes found to consistently mark early, ongoing and terminal maturational OA processes in patients. Furthermore, among the 37 OA risk genes that were significantly affected in cartilage by T3 were COL12A1, TNC, SPARC and PAPPA.
Conclusions: RNA-sequencing results show that metabolic activation and recuperation of growth plate morphology are induced by T3 in OA chondrocytes, indicating terminal maturation is accelerated. The molecular mechanisms involved in hypertrophy were linked to all stages of OA pathophysiology and will be used to validate disease models for drug testing.
Keywords: Cartilage; Chondrocyte; Hypertrophy; Lesioned; Osteochondral; Preserved; RNA-sequencing; T3; Terminal maturation.
© 2024. The Author(s).
Conflict of interest statement
The authors have declared no conflicts of interest.
Figures

References
-
- Vitaloni M, Botto-van Bemden A, Sciortino Contreras RM, Scotton D, Bibas M, Quintero M, et al. Global management of patients with knee osteoarthritis begins with quality of life assessment: a systematic review. BMC Musculoskeletal Disorders. 2019;20(1):493. doi: 10.1186/s12891-019-2895-3. - DOI - PMC - PubMed
-
- den Hollander W, Pulyakhina I, Boer C, Bomer N, van der Breggen R, Arindrarto W, et al. Annotating Transcriptional Effects of Genetic Variants in Disease-Relevant Tissue: Transcriptome-Wide Allelic Imbalance in Osteoarthritic Cartilage. Arthritis Rheumatol. 2019;71(4):561–70. doi: 10.1002/art.40748. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

