LINC complex alterations are a key feature of sporadic and familial ALS/FTD
- PMID: 38664831
- PMCID: PMC11046770
- DOI: 10.1186/s40478-024-01778-z
LINC complex alterations are a key feature of sporadic and familial ALS/FTD
Abstract
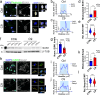
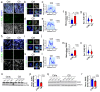
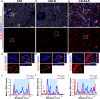
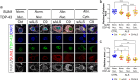
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder that primarily affects motor neurons, leading to progressive muscle weakness and loss of voluntary muscle control. While the exact cause of ALS is not fully understood, emerging research suggests that dysfunction of the nuclear envelope (NE) may contribute to disease pathogenesis and progression. The NE plays a role in ALS through several mechanisms, including nuclear pore defects, nucleocytoplasmic transport impairment, accumulation of mislocalized proteins, and nuclear morphology abnormalities. The LINC complex is the second biggest multi-protein complex in the NE and consists of the SUN1/2 proteins spanning the inner nuclear membrane and Nesprin proteins embedded in the outer membrane. The LINC complex, by interacting with both the nuclear lamina and the cytoskeleton, transmits mechanical forces to the nucleus regulating its morphology and functional homeostasis. In this study we show extensive alterations to the LINC complex in motor and cortical iPSC-derived neurons and spinal cord organoids carrying the ALS causative mutation in the C9ORF72 gene (C9). Importantly, we show that such alterations are present in vivo in a cohort of sporadic ALS and C9-ALS postmortem spinal cord and motor cortex specimens. We also found that LINC complex disruption strongly correlated with nuclear morphological alterations occurring in ALS neurons, independently of TDP43 mislocalization. Altogether, our data establish morphological and functional alterations to the LINC complex as important events in ALS pathogenic cascade, making this pathway a possible target for both biomarker and therapy development.
Keywords: ALS; C9ORF72; FTD; LINC complex; Nesprin; SUN.
© 2024. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures




Update of
-
LINC complex alterations are a hallmark of sporadic and familial ALS/FTD.bioRxiv [Preprint]. 2024 Mar 13:2024.03.08.584123. doi: 10.1101/2024.03.08.584123. bioRxiv. 2024. Update in: Acta Neuropathol Commun. 2024 Apr 25;12(1):69. doi: 10.1186/s40478-024-01778-z. PMID: 38559165 Free PMC article. Updated. Preprint.
References
-
- J.C., G. et al. The nuclear pore complex is compromised in ALS. Amyotroph Lateral Scler Front Degener (2016)
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

