Following the p63/Keratin5 basal cells in the sensory and non-sensory epithelia of the vomeronasal organ
- PMID: 38665067
- PMCID: PMC11141727
- DOI: 10.1002/dvg.23596
Following the p63/Keratin5 basal cells in the sensory and non-sensory epithelia of the vomeronasal organ
Abstract
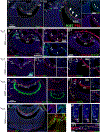
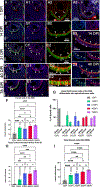
The vomeronasal organ (VNO) is a part of the accessory olfactory system, which detects pheromones and chemical factors that trigger a spectrum of sexual and social behaviors. The vomeronasal epithelium (VNE) shares several features with the epithelium of the main olfactory epithelium (MOE). However, it is a distinct neuroepithelium populated by chemosensory neurons that differ from the olfactory sensory neurons in cellular structure, receptor expression, and connectivity. The vomeronasal organ of rodents comprises a sensory epithelium (SE) and a thin non-sensory epithelium (NSE) that morphologically resembles the respiratory epithelium. Sox2-positive cells have been previously identified as the stem cell population that gives rise to neuronal progenitors in MOE and VNE. In addition, the MOE also comprises p63 positive horizontal basal cells, a second pool of quiescent stem cells that become active in response to injury. Immunolabeling against the transcription factor p63, Keratin-5 (Krt5), Krt14, NrCAM, and Krt5Cre tracing experiments highlighted the existence of horizontal basal cells distributed along the basal lamina of SE of the VNO. Single cell sequencing and genetic lineage tracing suggest that the vomeronasal horizontal basal cells arise from basal progenitors at the boundary between the SE and NSE proximal to the marginal zones. Moreover, our experiments revealed that the NSE of rodents is, like the respiratory epithelium, a stratified epithelium where the p63/Krt5+ basal progenitor cells self-replicate and give rise to the apical columnar cells facing the lumen of the VNO.
Keywords: Keratin5; basal progenitors; horizontal basal cells; non‐sensory epithelium; p63; stem cells; vomeronasal organ.
© 2024 Wiley Periodicals LLC.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

