Single-cell T cell receptor sequencing of paired human atherosclerotic plaques and blood reveals autoimmune-like features of expanded effector T cells
- PMID: 38665903
- PMCID: PMC11041750
- DOI: 10.1038/s44161-022-00208-4
Single-cell T cell receptor sequencing of paired human atherosclerotic plaques and blood reveals autoimmune-like features of expanded effector T cells
Abstract
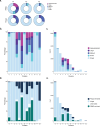
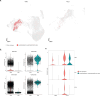
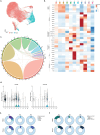
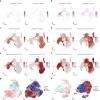
Atherosclerosis is a lipid-driven chronic inflammatory disease; however, whether it can be classified as an autoimmune disease remains unclear. In this study, we applied single-cell T cell receptor seqencing (scTCR-seq) on human carotid artery plaques and matched peripheral blood mononuclear cell samples to assess the extent of TCR clonality and antigen-specific activation within the various T cell subsets. We observed the highest degree of plaque-specific clonal expansion in effector CD4+ T cells, and these clonally expanded T cells expressed genes such as CD69, FOS and FOSB, indicative of recent TCR engagement, suggesting antigen-specific stimulation. CellChat analysis suggested multiple potential interactions of these effector CD4+ T cells with foam cells. Finally, we integrated a published scTCR-seq dataset of the autoimmune disease psoriatic arthritis, and we report various commonalities between the two diseases. In conclusion, our data suggest that atherosclerosis has an autoimmune compondent driven by autoreactive CD4+ T cells.
Keywords: Atherosclerosis; Autoimmunity.
© The Author(s) 2023.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures













Comment in
-
Understanding autoimmunity in atherosclerosis paves the way for novel therapies.Nat Cardiovasc Res. 2023 Mar;2(3):227-229. doi: 10.1038/s44161-023-00230-0. Nat Cardiovasc Res. 2023. PMID: 39196001 No abstract available.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
