Therapeutic efficacy of AAV-mediated restoration of PKP2 in arrhythmogenic cardiomyopathy
- PMID: 38665939
- PMCID: PMC11041734
- DOI: 10.1038/s44161-023-00378-9
Therapeutic efficacy of AAV-mediated restoration of PKP2 in arrhythmogenic cardiomyopathy
Abstract
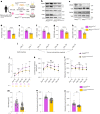
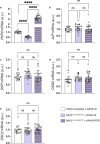
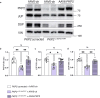
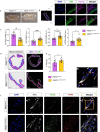
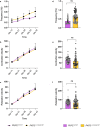
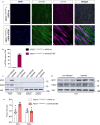
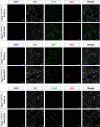
Arrhythmogenic cardiomyopathy is a severe cardiac disorder characterized by lethal arrhythmias and sudden cardiac death, with currently no effective treatment. Plakophilin 2 (PKP2) is the most frequently affected gene. Here we show that adeno-associated virus (AAV)-mediated delivery of PKP2 in PKP2c.2013delC/WT induced pluripotent stem cell-derived cardiomyocytes restored not only cardiac PKP2 levels but also the levels of other junctional proteins, found to be decreased in response to the mutation. PKP2 restoration improved sodium conduction, indicating rescue of the arrhythmic substrate in PKP2 mutant induced pluripotent stem cell-derived cardiomyocytes. Additionally, it enhanced contractile function and normalized contraction kinetics in PKP2 mutant engineered human myocardium. Recovery of desmosomal integrity and cardiac function was corroborated in vivo, by treating heterozygous Pkp2c.1755delA knock-in mice. Long-term treatment with AAV9-PKP2 prevented cardiac dysfunction in 12-month-old Pkp2c.1755delA/WT mice, without affecting wild-type mice. These findings encourage clinical exploration of PKP2 gene therapy for patients with PKP2 haploinsufficiency.
Keywords: Cardiomyopathies; Genetic vectors.
© The Author(s) 2023.
Conflict of interest statement
Competing interestsE.v.R. is a consultant for Tenaya Therapeutics and Novo Nordisk and is Chief Scientific Officer of Phlox Therapeutics. M.T., T.M. and W.H.Z. are inventors of patents related to the EHM technology and scientific advisors to myriamed GmbH. W.H.Z. is founder and shareholder of myriamed GmbH. M.G. is founder, consultant, member of the Board and equity holder in Purespring Therapeutics, Forcefield Therapeutics and Heqet Therapeutics. All other authors declare that they have no competing interests.
Figures








Comment in
-
Restoring PKP2 in arrhythmogenic cardiomyopathy.Nat Cardiovasc Res. 2023 Dec;2(12):1115-1116. doi: 10.1038/s44161-023-00382-z. Nat Cardiovasc Res. 2023. PMID: 39196151 No abstract available.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
