Flow cytometric immunophenotypic differentiation patterns of bone marrow eosinophilopoiesis
- PMID: 38666394
- PMCID: PMC11442120
- DOI: 10.1002/cyto.b.22174
Flow cytometric immunophenotypic differentiation patterns of bone marrow eosinophilopoiesis
Abstract
Background: Flow cytometry has been widely used to study immunophenotypic patterns of maturation of most hematopoietic lineages in normal human bone marrow aspirates, thus allowing identification of changes in patterns in many myeloid malignancies. Eosinophils play an important role in a wide variety of disorders, including some myeloid neoplasms. However, changes in flow cytometric immunophenotypic patterns during normal and abnormal bone marrow eosinophilopoiesis have not been well studied.
Methods: Fresh bone marrow aspirates from 15 healthy donors, 19 patients with hypereosinophilic syndromes (HES), and 11 patients with systemic mastocytosis (SM) were analyzed for candidate markers that included EMR-1, Siglec-8, CCR3, CD9, CD11a, CD11b, CD11c, CD13, CD16, CD29, CD34, CD38, CD45, CD44, CD49d, CD49f, CD54, CD62L, CD69, CD117, CD125 (IL-5Rα), HLA-DR, using 10 parameter flow cytometry. Putative CD34-negative immature and mature normal eosinophil populations were first identified based on changes in expression of the above markers in healthy donors, then confirmed using fluorescence-based cell sorting and morphological evaluation of cytospin preparations. The normal immunophenotypic patterns were then compared to immunophenotypic patterns of eosinophilopoiesis in patients with HES and SM.
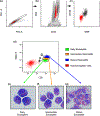
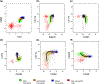
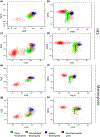
Results: The eosinophilic lineage was first verified using the human eosinophil-specific antibody EMR-1 in combination with anti-IL-5Rα antibody. Then, a combination of Siglec-8, CD9, CD11b, CCR3, CD49d, and CD49f antibodies was used to delineate normal eosinophilic maturational patterns. Early stages (eosinophilic promyelocytes/myelocytes) were identified as Siglec-8 dim/CD11b dim to moderate/CD9 dim/CCR3 dim/CD49d bright/CD49f dim, intermediate stages (eosinophilic myelocytes/metamyelocytes) as Siglec-8 moderate/CD11b moderate to bright/CD9 moderate/CCR3 moderate/CD49d moderate/CD49f moderate and mature bands/segmented eosinophils as Siglec-8 bright/CD11b bright/CD9 bright/CCR3 bright/CD49d dim/CD49f bright. Overall maturational patterns were also similar in patients with HES and SM; however, the expression levels of several surface markers were altered compared to normal eosinophils.
Conclusion: A novel flow cytometric antibody panel was devised to detect alterations in immunophenotypic patterns of bone marrow eosinophil maturation and evaluated in normal, HES and SM samples. This approach will allow us to elucidate changes in immunophenotypic patterns of bone marrow eosinophilopoiesis in other hematological diseases.
Keywords: adhesion molecules; bone marrow; eosinophil maturation; flow cytometry; hematological disease.
Published 2024. This article is a U.S. Government work and is in the public domain in the USA. Cytometry Part B: Clinical Cytometry published by Wiley Periodicals LLC on behalf of International Clinical Cytometry Society.
Figures



References
-
- Al-Rabia MW, Blaylock MG, Sexton DW, Thomson L, & Walsh GM (2003). Granule protein changes and membrane receptor phenotype in maturing human eosinophils cultured from CD34+ progenitors. Clinical and Experimental Allergy, 33, 640–648. - PubMed
-
- Bainton DF (1992). Inflammation: Basic principles and clinical correlates. In Goldstein R, Gallin JI, & Snyderman R (Eds.), Developmental biology of neutrophils and eosinophils. Raven Press.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

