Occurrence of Antimicrobial-Resistant Bacteria in Intestinal Contents of Wild Marine Fish in Chile
- PMID: 38667008
- PMCID: PMC11047320
- DOI: 10.3390/antibiotics13040332
Occurrence of Antimicrobial-Resistant Bacteria in Intestinal Contents of Wild Marine Fish in Chile
Abstract
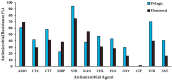
Antimicrobial-resistant bacteria (ARB) from the intestinal contents of wild fish may have a relevant ecological significance and could be used as indicators of antimicrobial-resistance dissemination in natural bacterial populations in water bodies impacted by urban contamination. Thus, the occurrence of ARB in the intestinal contents of pelagic and demersal wild fishes captured in anthropogenic-impacted Coquimbo Bay in Chile was studied. Culturable counts of total and antimicrobial-resistant bacteria were determined by a spread plate method using Trypticase soy agar and R2A media, both alone and supplemented with the antimicrobials amoxicillin, streptomycin, florfenicol, oxytetracycline and ciprofloxacin, respectively. Heterotrophic plate counts of pelagic and demersal fishes ranged from 1.72 × 106 CFU g-1 to 3.62 × 109 CFU g-1, showing variable proportions of antimicrobial resistance. Representative antimicrobial-resistant isolates were identified by 16S rRNA gene sequencing, and isolates (74) from pelagic fishes mainly belonged to Pseudomonas (50.0%) and Shewanella (17.6%) genera, whereas isolates (68) from demersal fishes mainly belonged to Vibrio (33.8%) and Pseudomonas (26.5%) genera. Antimicrobial-resistant isolates were tested for susceptibility to 12 antimicrobials by an agar disk diffusion method, showing highest resistance to streptomycin (85.2%) and amoxicillin (64.8%), and lowest resistance to oxytetracycline (23.2%) and ciprofloxacin (0.7%). Only furazolidone and trimethoprim/sulfamethoxazole were statistically different (p < 0.05) in comparisons between isolates from pelagic and demersal wild fishes. Furthermore, an important number of these isolates carried plasmids (53.5%) and produced Extended-Spectrum-β-lactamases (ESBL) (16.9%), whereas the detection of Metallo-β-Lactamases and class 1-integron was rare. This study provides evidence that wild fish are important reservoirs and spreading-vehicles of ARB, carrying plasmids and producing ESBLs in Chilean marine environments.
Keywords: Chile; antibacterial resistant bacteria; antimicrobial resistance; beta lactamases; fish bacteriology; marine fishes.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Fu J., Yang D., Jin M., Liu W., Zhao X., Li C., Zhao T., Wang J., Gao Z., Shen Z. Aquatic animals promote antibiotic resistance gene dissemination in water via conjugation: Role of different regions within the zebra fish intestinal tract, and impact on fish intestinal microbiota. Mol. Ecol. 2017;26:5318–5333. doi: 10.1111/mec.14255. - DOI - PubMed
-
- Miranda C.D. Antimicrobial resistance in salmonid farming. In: Keen P.L., Montforts M.H.M.M., editors. Antimicrobial Resistance in the Environment. Wiley-Blackwell; Hoboken, NJ, USA: 2012. pp. 423–451. Chapter 22.
-
- Domínguez M., Miranda C.D., Fuentes O., de la Fuente M., Godoy F.A., Bello-Toledo H., González-Rocha G. Occurrence of transferable integrons and sul and dfr genes among sulfonamide-and/or trimethoprim-resistant bacteria isolated from Chilean salmonid farms. Front. Microbiol. 2019;10:748. doi: 10.3389/fmicb.2019.00748. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

