Integrated Omics Analysis Reveals Key Pathways in Cotton Defense against Mirid Bug (Adelphocoris suturalis Jakovlev) Feeding
- PMID: 38667384
- PMCID: PMC11049813
- DOI: 10.3390/insects15040254
Integrated Omics Analysis Reveals Key Pathways in Cotton Defense against Mirid Bug (Adelphocoris suturalis Jakovlev) Feeding
Abstract
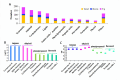
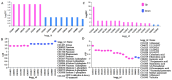
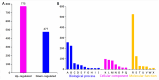
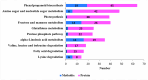
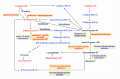
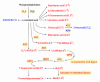
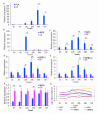
The recent dominance of Adelphocoris suturalis Jakovlev as the primary cotton field pest in Bt-cotton-cultivated areas has generated significant interest in cotton pest control research. This study addresses the limited understanding of cotton defense mechanisms triggered by A. suturalis feeding. Utilizing LC-QTOF-MS, we analyzed cotton metabolomic changes induced by A. suturalis, and identified 496 differential positive ions (374 upregulated, 122 downregulated) across 11 categories, such as terpenoids, alkaloids, phenylpropanoids, flavonoids, isoflavones, etc. Subsequent iTRAQ-LC-MS/MS analysis of the cotton proteome revealed 1569 differential proteins enriched in 35 metabolic pathways. Integrated metabolome and proteome analysis highlighted significant upregulation of 17 (89%) proteases in the α-linolenic acid (ALA) metabolism pathway, concomitant with a significant increase in 14 (88%) associated metabolites. Conversely, 19 (73%) proteases in the fructose and mannose biosynthesis pathway were downregulated, with 7 (27%) upregulated proteases corresponding to the downregulation of 8 pathway-associated metabolites. Expression analysis of key regulators in the ALA pathway, including allene oxidase synthase (AOS), phospholipase A (PLA), allene oxidative cyclase (AOC), and 12-oxophytodienoate reductase3 (OPR3), demonstrated significant responses to A. suturalis feeding. Finally, this study pioneers the exploration of molecular mechanisms in the plant-insect relationship, thereby offering insights into potential novel control strategies against this cotton pest.
Keywords: Adelphocoris suturalis Jakovlev; cotton plant defense; fructose and mannose biosynthesis pathway; key regulators; α-linolenic acid metabolism pathway.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Xu T., Zhou Q., Chen W., Zhang G., He G.F., Gu D.X., Zhang W.Q. Involvement of Jasmonate-signaling pathway in the herbivore-induced rice plant defense. Chin. Sci. Bull. 2003;48:1982–1987. doi: 10.1007/BF03183991. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

