KRAS: Biology, Inhibition, and Mechanisms of Inhibitor Resistance
- PMID: 38668053
- PMCID: PMC11049385
- DOI: 10.3390/curroncol31040150
KRAS: Biology, Inhibition, and Mechanisms of Inhibitor Resistance
Abstract
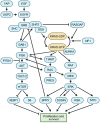
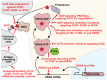
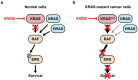
KRAS is a small GTPase that is among the most commonly mutated oncogenes in cancer. Here, we discuss KRAS biology, therapeutic avenues to target it, and mechanisms of resistance that tumors employ in response to KRAS inhibition. Several strategies are under investigation for inhibiting oncogenic KRAS, including small molecule compounds targeting specific KRAS mutations, pan-KRAS inhibitors, PROTACs, siRNAs, PNAs, and mutant KRAS-specific immunostimulatory strategies. A central challenge to therapeutic effectiveness is the frequent development of resistance to these treatments. Direct resistance mechanisms can involve KRAS mutations that reduce drug efficacy or copy number alterations that increase the expression of mutant KRAS. Indirect resistance mechanisms arise from mutations that can rescue mutant KRAS-dependent cells either by reactivating the same signaling or via alternative pathways. Further, non-mutational forms of resistance can take the form of epigenetic marks, transcriptional reprogramming, or alterations within the tumor microenvironment. As the possible strategies to inhibit KRAS expand, understanding the nuances of resistance mechanisms is paramount to the development of both enhanced therapeutics and innovative drug combinations.
Keywords: KRAS; cancer; immunotherapy; inhibitors; mutation; oncogene; resistance; targeted therapy.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

