Clinical Strains of Mycobacterium tuberculosis Representing Different Genotype Families Exhibit Distinct Propensities to Adopt the Differentially Culturable State
- PMID: 38668273
- PMCID: PMC11054447
- DOI: 10.3390/pathogens13040318
Clinical Strains of Mycobacterium tuberculosis Representing Different Genotype Families Exhibit Distinct Propensities to Adopt the Differentially Culturable State
Abstract
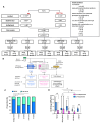
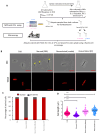
Growing evidence points to the presence of differentially culturable tubercle bacteria (DCTB) in clinical specimens from individuals with active tuberculosis (TB) disease. These bacteria are unable to grow on solid media but can resuscitate in liquid media. Given the epidemiological success of certain clinical genotype families of Mycobacterium tuberculosis, we hypothesize that different strains may have distinct mechanisms of adaptation and tolerance. We used an in vitro carbon starvation model to determine the propensity of strains from lineages 2 and 4 that included the Beijing and LAM families respectively, to generate DCTB. Beijing strains were associated with a greater propensity to produce DCTB compared to LAM strains. Furthermore, LAM strains required culture filtrate (CF) for resuscitation whilst starved Beijing strains were not dependent on CF. Moreover, Beijing strains showed improved resuscitation with cognate CF, suggesting the presence of unique growth stimulatory molecules in this family. Analysis of starved Beijing and LAM strains showed longer cells, which with resuscitation were restored to a shorter length. Cell wall staining with fluorescent D-amino acids identified strain-specific incorporation patterns, indicating that cell surface remodeling during resuscitation was distinct between clinical strains. Collectively, our data demonstrate that M. tuberculosis clinical strains from different genotype lineages have differential propensities to generate DCTB, which may have implications for TB treatment success.
Keywords: Beijing strains; LAM strains; Mycobacterium tuberculosis; differentially culturable tubercle bacteria; tuberculosis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Chengalroyen M.D., Beukes G.M., Gordhan B.G., Streicher E.M., Churchyard G., Hafner R., Warren R., Otwombe K., Martinson N., Kana B.D. Detection and quantification of differentially culturable tubercle bacteria in sputum from patients with tuberculosis. Am. J. Respir. Crit. Care Med. 2016;194:1532–1540. doi: 10.1164/rccm.201604-0769OC. - DOI - PMC - PubMed
-
- Gordhan B.G., Sewcharran A., Letsoalo M., Chinappa T., Yende-Zuma N., Padayatchi N., Naidoo K., Kana B.D. Detection of differentially culturable tubercle bacteria in sputum from drug-resistant tuberculosis patients. Front. Cell. Infect. Microbiol. 2022;12:949370. doi: 10.3389/fcimb.2022.949370. - DOI - PMC - PubMed
-
- Peters J.S., McIvor A., Papadopoulos A.O., Masangana T., Gordhan B.G., Waja Z., Otwombe K., Letutu M., Kamariza M., Sterling T.R., et al. Differentially culturable tubercle bacteria as a measure of tuberculosis treatment response. Front. Cell. Infect. Microbiol. 2022;12:1064148. doi: 10.3389/fcimb.2022.1064148. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

