Generation of rat forebrain tissues in mice
- PMID: 38670071
- PMCID: PMC11646705
- DOI: 10.1016/j.cell.2024.03.017
Generation of rat forebrain tissues in mice
Abstract
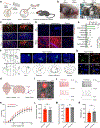
Interspecies blastocyst complementation (IBC) provides a unique platform to study development and holds the potential to overcome worldwide organ shortages. Despite recent successes, brain tissue has not been achieved through IBC. Here, we developed an optimized IBC strategy based on C-CRISPR, which facilitated rapid screening of candidate genes and identified that Hesx1 deficiency supported the generation of rat forebrain tissue in mice via IBC. Xenogeneic rat forebrain tissues in adult mice were structurally and functionally intact. Cross-species comparative analyses revealed that rat forebrain tissues developed at the same pace as the mouse host but maintained rat-like transcriptome profiles. The chimeric rate of rat cells gradually decreased as development progressed, suggesting xenogeneic barriers during mid-to-late pre-natal development. Interspecies forebrain complementation opens the door for studying evolutionarily conserved and divergent mechanisms underlying brain development and cognitive function. The C-CRISPR-based IBC strategy holds great potential to broaden the study and application of interspecies organogenesis.
Keywords: C-CRISPR; interspecies blastocyst complementation; interspecies chimeras; interspecies forebrain blastocyst complementation; interspecies neural blastocyst complementation; interspecies organogenesis; rat-mouse chimeras.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

