Functional sensory circuits built from neurons of two species
- PMID: 38670072
- PMCID: PMC11293795
- DOI: 10.1016/j.cell.2024.03.042
Functional sensory circuits built from neurons of two species
Abstract
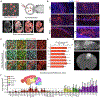
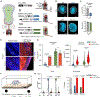
A central question for regenerative neuroscience is whether synthetic neural circuits, such as those built from two species, can function in an intact brain. Here, we apply blastocyst complementation to selectively build and test interspecies neural circuits. Despite approximately 10-20 million years of evolution, and prominent species differences in brain size, rat pluripotent stem cells injected into mouse blastocysts develop and persist throughout the mouse brain. Unexpectedly, the mouse niche reprograms the birth dates of rat neurons in the cortex and hippocampus, supporting rat-mouse synaptic activity. When mouse olfactory neurons are genetically silenced or killed, rat neurons restore information flow to odor processing circuits. Moreover, they rescue the primal behavior of food seeking, although less well than mouse neurons. By revealing that a mouse can sense the world using neurons from another species, we establish neural blastocyst complementation as a powerful tool to identify conserved mechanisms of brain development, plasticity, and repair.
Keywords: blastocyst complementation; brain evolution; brain regeneration; development; disease models; interspecies chimeras; neural circuits; neurons; olfaction; pluripotent stem cells.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests K.K.B. is on the SAB of Gameto Therapeutics. B.T.T. is a current employee of Janssen Research and Development. P.O. is a founder of Theracast. J.C.I.B is a founding scientist and director of the San Diego Institute of Science, Altos Labs.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

